Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) is one of the most important animal pathogens in swine industry worldwide. In this study, we isolated the large-plaque forming variant virus designated PL97-1/LP1 from the parental strain PL97-1, the first Korean strain of PRRSV, isolated from the serum of an infected pig in 1997. We found that the 15411-nucleotide genome of PL97-1/LP1 consisted of a 189-nucleotide 5′ untranslated region (UTR), a 15071-nucleotide protein-coding region, and a 151-nucleotide 3'UTR, followed by a poly (A) tail of about 50∼60 nucleotides in size. The 5′-end of PL97-1/LP1 began with ATGACGTAT. Comparison of the PL97-1/LP1 genome with the 11 fully sequenced PRRSV genomes currently available revealed sequence similarity from 99.6∼99.7% (the North American VR-2332 and two VR-2332-derived vaccine strains MLV RespPRRS/Repro and RespPRRS MLV) to 62.0% (the Dutch Lelystad strain). Phylogenetc analysis revealed that PL97-1/LP1 is most closely related to the North American genotype VR-2332, two VR-2332-derived vaccine strains, and Chinese BJ-4. It is distantly related to the European genotype Lelystad. The entire nucleotide sequence of PL97-1/LP1 was identical to that of the parental virus PL97–1 except for three silent nucleotide substitutions, one in ORF1a (U4230C), one in ORF1b (C10977U), and one in ORF5 (U13976A). This nucleotide sequence has been submitted to the GenBank database under the accession number AY612613.
Go to : 
References
1). Allende R, Lewis TL, Lu Z, Rock DL, Kutish GF, Ali A, Doster AR, Osorio FA. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. J Gen Virol. 80:307–315. 1999.

2). Andreyev VG, Wesley RD, Mengeling WL, Vorwald AC, Lager KM. Genetic variation and phylogenetic relationships of 22 porcine reproductive and respiratory syndrome virus (PRRSV) field strains based on sequence analysis of open reading frame 5. Arch Virol. 142:993–1001. 1997.

3). Benfield DA, Nelson E, Collins JE, Harris L, Goyal SM, Robison D, Christianson WT, Morrison RB, Gorcyca D, Chladek D. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J Vet Diagn Invest. 4:127–133. 1992.

4). Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 57:537–547. 1989.

5). Cavanagh D. Nidovirales: a new order comprising Corona-viridae and Arteriviridae. Arch Virol. 142:629–633. 1997.
6). Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, Goyal SM, McCullough S, Morrison RB, Joo HS, Gorcyca DE, Chladek DW. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest. 4:117–126. 1992.

7). Conzelmann KK, Visser N, Van Woensel P, Thiel HJ. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group. Virology. 193:329–339. 1993.

8). Gagnon CA, Dea S. Differentiation between porcine reproductive and respiratory syndrome virus isolates by restriction fragment length polymorphism of their ORFs 6 and 7 genes. Can J Vet Res. 62:110–116. 1998.
9). Gonin P, Pirzadeh B, Gagnon CA, Dea S. Seroneutralization of porcine reproductive and respiratory syndrome virus correlates with antibody response to the GP5 major envelope glycoprotein. J Vet Diagn Invest. 11:20–26. 1999.

10). Halbur PG, Paul PS, Frey ML, Landgraf J, Eernisse K, Meng XJ, Lum MA, Andrews JJ, Rathje JA. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol. 32:648–660. 1995.

11). Halbur PG, Paul PS, Meng XJ, Lum MA, Andrews JJ, Rathje JA. Comparative pathogenicity of nine US porcine reproductive and respiratory syndrome virus (PRRSV) isolates in a five-week-old cesarean-derived, colostrum-deprived pig model. J Vet Diagn Invest. 8:11–20. 1996.

12). Kang SY, Yun SI, Park HS, Park CK, Choi HS, Lee YM. Molecular characterization of PL97–1, the first Korean isolate of the porcine reproductive and respiratory syndrome virus. Virus Res. 104:165–179. 2004.

13). Kapur V, Elam MR, Pawlovich TM, Murtaugh MP. Genetic variation in porcine reproductive and respiratory syndrome virus isolates in the midwestern United States. J Gen Virol. 77:1271–1276. 1996.

14). Keffaber KK. Reproductive failure of unknown etiology. Am Assoc Swine Pract Newslett. 1:1–9. 1989.
15). Kwang J, Kim HS, Joo HS. Cloning, expression, and sequence analysis of the ORF4 gene of the porcine reproductive and respiratory syndrome virus MN-1b. J Vet Diagn Invest. 6:293–296. 1994.

16). Madsen KG, Hansen CM, Madsen ES, Strandbygaard B, Botner A, Sorensen KJ. Sequence analysis of porcine reproductive and respiratory syndrome virus of the American type collected from Danish swine herds. Arch Virol. 143:1683–1700. 1998.

17). Mardassi H, Mounir S, Dea S. Identification of major differences in the nucleocapsid protein genes of a Quebec strain and European strains of porcine reproductive and respiratory syndrome virus. J Gen Virol. 75:681–685. 1994.

18). Meng XJ, Paul PS, Halbur PG. Molecular cloning and nucleotide sequencing of the 3-terminal genomic RNA of the porcine reproductive and respiratory syndrome virus. J Gen Virol. 75:1795–1801. 1994.

19). Meng XJ, Paul PS, Halbur PG, Lum MA. Phylogenetic analyses of the putative M (ORF 6) and N (ORF 7) genes of porcine reproductive and respiratory syndrome virus (PRRSV): implication for the existence of two genotypes of PRRSV in the USA and Europe. Arch Virol. 140:745–755. 1995.

20). Meulenberg JJM, Hulst MM, de Meijer EJ, Moonen PLJM, den Besten A, de Kluyver EP, Wensvoort G, Moormann RJM. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 192:62–72. 1993.

21). Morozov I, Meng XJ, Paul PS. Sequence analysis of open reading frames (ORFs) 2–4 of a US isolate of porcine reproductive and respiratory syndrome virus. Arch Virol. 140:1313–1319. 1995.
22). Murtaugh MP, Elam MR, Kakach LT. Comparison of the structural protein coding sequences of the VR-2332 and Lelystad virus strains of the PRRS virus. Arch Viro. 140:1451–1460. 1995.

23). Murtaugh MP, Faaberg KS, Laber J, Elam M, Kapur V. Genetic variation in the PRRS virus. Adv Exp Med Biol. 440:787–794. 1998.

24). Nelsen CJ, Murtaugh MP, Faaberg KS. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol. 73:270–280. 1999.

25). Oleksiewicz MB, Botner A, Nielsen J, Storgaard T. Determination of 5-leader sequences from radically disparate strains of porcine reproductive and respiratory syndrome virus reveals the presence of highly conserved sequence motifs. Arch Virol. 144:981–987. 1999.

26). Page RDM. Treeview: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 12:357–358. 1996.
27). Paton DJ, Brown IH, Edwards S, Wensvoort G. ‘Blue ear’ disease of pigs. Vet Rec. 128:617. 1991.

28). Rowland RR, Steffen M, Ackerman T, Benfield DA. The evolution of porcine reproductive and respiratory syndrome virus: quasispecies and emergence of a virus subpopulation during infection of pigs with VR-2332. Virology. 259:262–266. 1999.

29). Saitou N, Nei M. The Neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425. 1987.
30). Salles FJ, Strickland S. Rapid and sensitive analysis of mRNA polyadenylation states by PCR. PCR Methods Appl. 4:317–321. 1995.

31). Snijder EJ, Meulenberg JJM. The molecular biology of arteriviruses. J Gen Virol. 79:961–979. 1998.

32). Snijder EJ, Meulenberg JJM. Arteriviruses. Knipe D.M., Howley P.M, editors. (Eds.),. Fields Virology. fourth ed.vol.1:Lippincott Williams and Wilkins;PA: pp.p. 1205–1220. 2001.
33). Snijder EJ, van Tol H, Pedersen KW, Raamsman MJ, de Vries AA. Identification of a novel structural protein of arteri-viruses. J Virol. 73:6335–6345. 1999.

34). Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality tools. Nucleic Acids Res. 25:4876–4882. 1997.
35). Ward CD, Stokes MA, Flanegan JB. Direct measurement of the poliovirus RNA polymerase error frequency in vitro. J Virol. 62:558–562. 1988.

36). Wensvoort G, Terpstra C, Pol JM, ter Laak EA, Bloemraad M, de Kluyver EP, Kragten C, van Buiten L, den Besten A, Wagenaar F, Broekhuijsen JM, Moonen PLJM, Zetstra T, de Boer EA, Tibben HJ, de Jong MF, vant Veld P, Groenland GJR, van Gennep JA, Voets MT, Verheijden JHM, Braamskamp J. Mystery swine disease in the Netherlands: the isolation of Lelystad virus. Vet Q. 13:121–130. 1991.

Go to : 
 | Figure 1.Synthesis of cDNA amplicons encompassing the entire genomic RNA of the PRRSV PL97-1/LP1 strain. The full-length viral genomic RNA is schematically drawn at the top and the four overlapping cDNA amplicons are shown below. PF1, nt 180-5297 PF2, nt 3708-9108 PF3, nt 7688-13051 PF4, nt 9610–15238. |
 | Figure 2.Synthesis of cDNA amplicons encompassing the entire genomic RNA of the PRRSV PL97–1 strain. (A) Synthesis by long RT-PCR of four long overlapping cDNA amplicons (PF1, PF2, PF3, and PF4) that cover the full-length viral genomic RNA except for the 5′ and 3′ termini. Four PRRSV-specific overlapping cDNA amplicons are synthesized in the presence of an RNaseH(–) RT (lanes 1, 3, 5, and 7) but not in its absence (lanes 2, 4, 6, and 8). The products were analyzed on a 1% agarose gel and visualized by staining with ethidium bromide. Lanes 1-2, PF1 amplicons; lanes 3-4, PF2 amplicons; lane 5-6, PF3 amplicons, lanes 7-8, PF4 amplicons. M, 1 kb DNA marker indicated in base pairs. (B) Synthesis of PRRSV-specific amplicons by a modified 5'RACE protocol from the OligoT-ligated PRRSV cDNAs to determine the 5′-terminal sequence. RT reaction used to synthesize the first-strand cDNA corresponding to the 5′-end portion of the genome was conducted in the presence (lanes 1-2) or absence (lane 3) of RNaseH(–) RT. The OligoT was ligated to the 3′-end of first-strand cDNA using T4 RNA ligase in the presence (lanes 1 and 3) or absence (lane 2) of OligoT. The OligoT-ligated first-strand cDNA was then PCR-amplified using a pair of primers. PRRSV-specific cDNA amplicons were analyzed on a 1.5% agarose gel and visualized by staining with ethidium bromide. M, 100 bp DNA ladder indicated in base pairs. (C) Synthesis of PRRSV-specific amplicons from the OligoT-ligated PRRSV genomic RNAs by a modified 3'RACE protocol to determine the 3′-terminal sequence. The OligoT was ligated to the 3′-end of viral genomic RNA using T4 RNA ligase in the presence (lanes 1 and 3) or absence (lane 2) of OligoT. The OligoT-ligated viral RNA was then used for cDNA synthesis in the presence (lanes 1-2) or absence (lane 3) of RNaseH(–) RT. After PCR amplification, PRRSV-specific cDNA amplicons were analyzed on a 1.5% agarose gel and visualized by staining with ethidium bromide. M, 100 bp DNA ladder indicated in base pairs. |
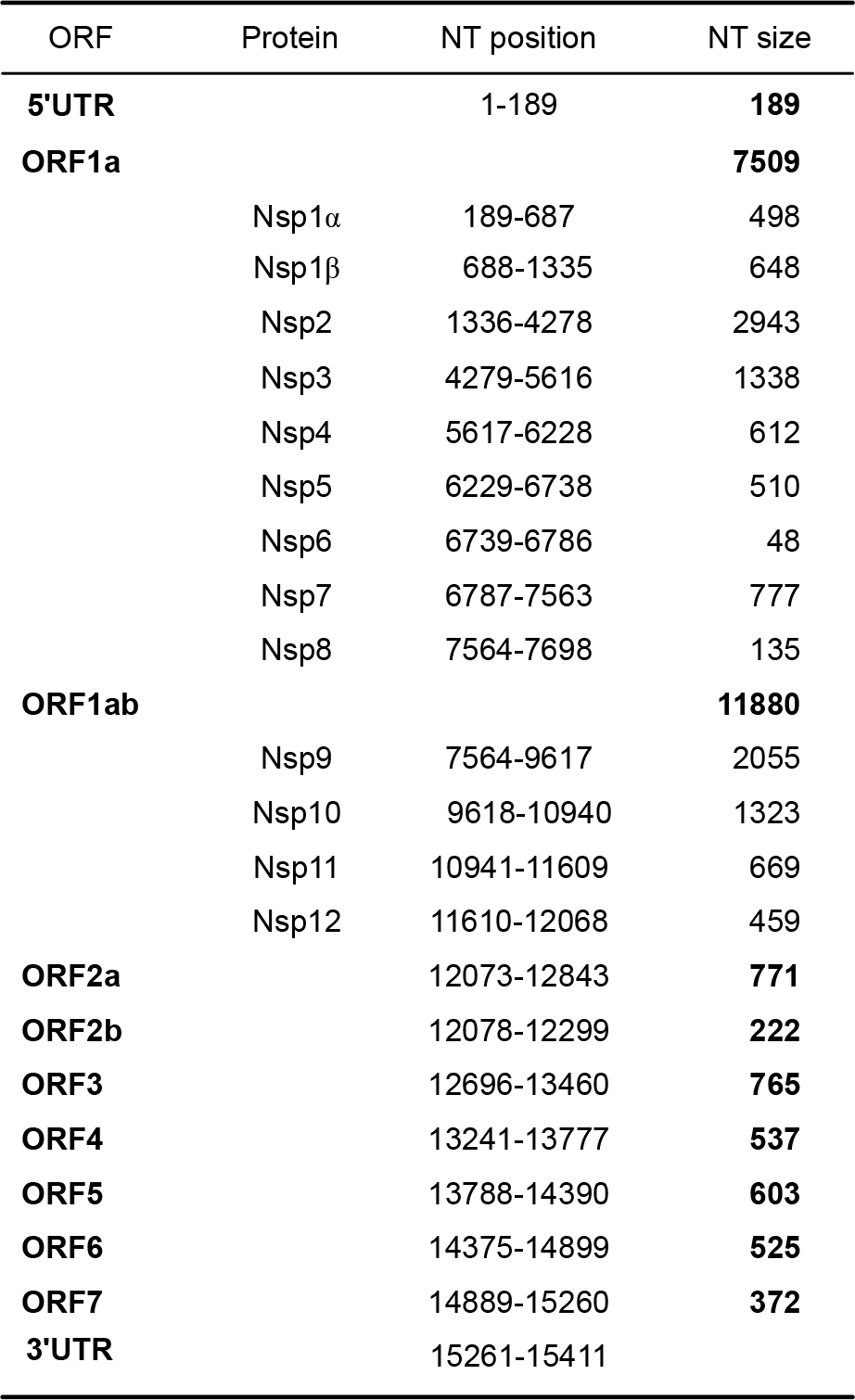 | Figure 3.The genome organization and the predicted polypeptides encoded by the PRRSV PL97–1/LP1 viral genome RNA. UTR, untranslated region; ORF, open reading frame; Nsp, non-structural protein; NT, nucleotide. |
 | Figure 4.Pairwise comparisons of full-length genome of PRRSV isolates. Percentage identity of nucleotide and amino acid sequences are shown above and below the diagonal line, respectively. |
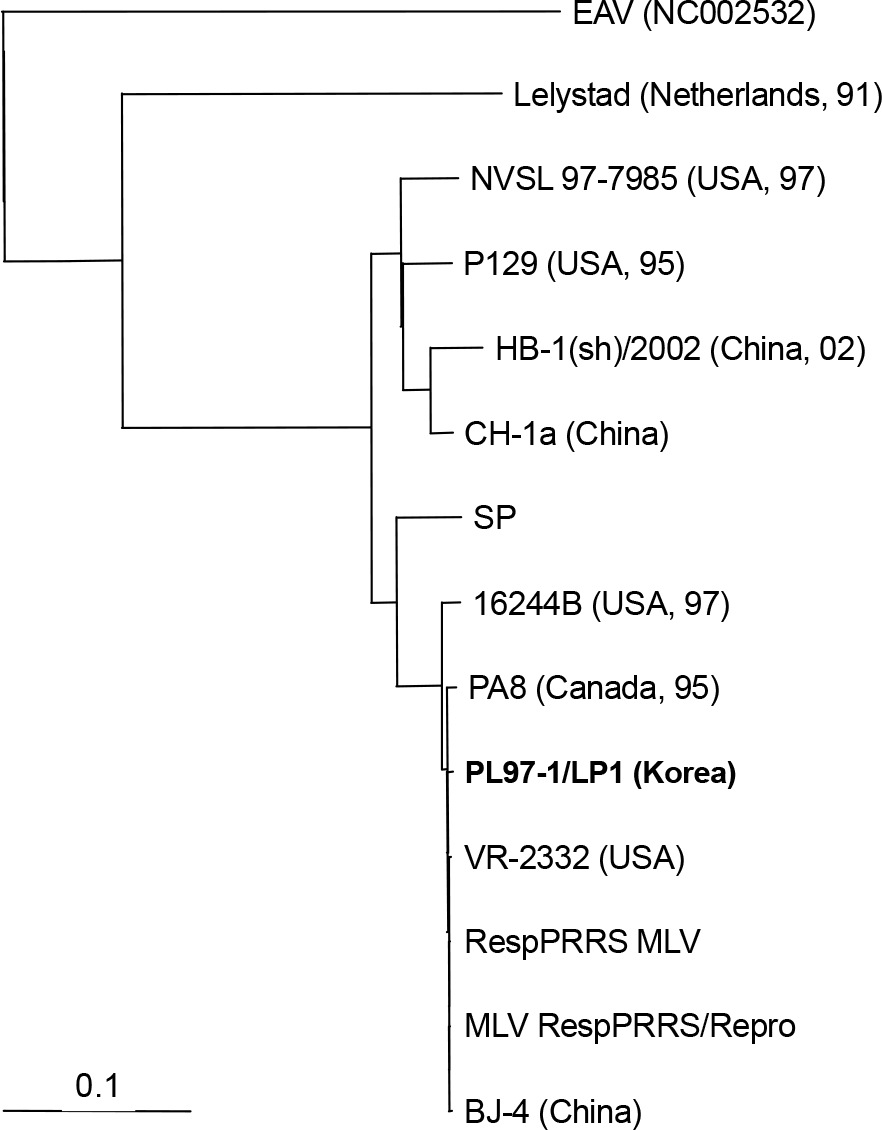 | Figure 5.A phylogenetic tree constructed with the nucleotide sequence of the full-length genome of all 12 available PRRSV isolates. The phylogenetic tree was constructed using the neighborjoining method in ClustralX method. The scale bar at the bottom of the tree represent the number of nucleotide substitutions per site. The tree was rooted using the nucleotide sequence of EAV, a member of the Arteriviridae family. The strain name is followed by the country and the year of isolation in two digits. |
 | Figure 6.Predicted pseudoknot structure of the ribosomal frame-shift region of PRRSV PL97–1/LP1. The heptanucleotide sequence (UUUAAAC) slippery sequence is in a box. The stop codon UAG of ORF1a is in the boldface type. Numbers indicate nucleotide position. |
Table 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea | Postitionb | Polarity | Purpose |
|---|---|---|---|---|
| PR1RT | 5′-TAGGATGGTGAGGGGGTG | 5332∼5349 | – | RT |
| PR1F | 5′-CCCTTTAACCATGTCT | 180∼195 | + | PCR |
| PR1R | 5′-CAAAGCAACCAGGTAA | 5282∼5297 | – | PCR |
| PR2RT | 5′-GAGCATGTCCTCAAACTT | 9168∼9185 | – | RT |
| PR2F | 5′-CCGGATATCGTCGCGG | 3708∼3723 | + | PCR |
| PR2R | 5′-CCATATGCTGTGCATA | 9093∼9108 | – | PCR |
| PR3RT | 5′-CACATTCCCTATCCCGAA | 13074∼13091 | – | RT |
| PR3F | 5′-GTTTAAACTGCTAGCC | 7688∼7703 | + | PCR |
| PR3R | 5′-GTGTAGCTGAAGGACA | 13036∼13051 | – | PCR |
| PR4RT | 5′-CTAATTGAATAGGTGACT | 15342∼15359 | – | RT |
| PR4F | 5′-ATTATGAGGGGAAGAA | 9610∼9625 | + | PCR |
| PR4R | 5′-ACGCGGATCAGGCGCA | 15223∼15238 | – | PCR |
| PR41 | 5′-GGAGAAGCCCCATTTTCC | 15038∼15055 | + | PCR |
| PR49 | 5′-CGACCCGTACCATTCTTT | 476∼493 | – | PCR |
| PR50 | 5′-AAAAGTCTTCAGGCTTGG | 692∼709 | – | RT |
| OligoT | 5′-CCAGTGTTGTGGCCTGCAGGGCGAATT | RNA ligation | ||
| OligoTR | 5′-GATGAATTCGCCCTGCAGGCCACAACA | RT, PCR |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download