Abstract
A number of recombinant proteins isolated from cell sources are being produced for biopharmaceuticals. Although most biopharmaceuticals are highly purified, there is a safety concern that such recombinant products could be contaminated with impurities including adventitious virus, mycoplasma, endotoxin and oncogenic DNA. Residual DNA in recombinant biopharmaceuticals is a potential risk factor and must be evaluated and removed to meet the regulatory guidelines. Recombinant HPV type 16 L1 VLPs, recombinant protein produced in Spodoptera frugiperda (Sf) 9 insect cells, is a HPV subunit vaccine candidate which has been studied as a preventive vaccine of cervical cancers. In this study, we performed detection and quantification of residual cellular DNA in the production of recombinant HPV type 16 L1 VLPs. HPV-16 L1 VLPs were purified by processes including detergent lysis, sonication treatment, sucrose cushion centrifugation, CsCl equilibrium density centrifugation, and DNase treatment which was added to inactivate residual cellular DNA after CsCl centrifugation step. We have developed a precise assay based on a dot-blot hybridization using digoxigenin random primed labeling DNA probes for the detection and quantification of residual cellular DNA during the purification process and final products. Detection limit of residual cellular DNA was 0.1 ng in this assay and the amount of residual cellular DNA in the final product was 0.5 ng∼1 ng per 100 μg of protein. This study describes safer and more sensitive methods alternative to radioactive techniques employed for residual cellular DNA quantification of biopharmaceuticals produced by recombinant protein technology and presents method validation data demonstrating precision and reproducibility.
Go to : 
References
1). Bosch FX, Manos MM, Munoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shah KV. Prevalence of human papillomavirus in cervical cancer: A worldwide prospective. J Natl Cancer Inst. 87:796–802. 1995.
2). Briggs J, Panfili PR. Quantitation of DNA and protein impurities in biopharmaceuticals. Anal Chem. 63(9):850–859. 1991.

3). Diane M Harper ELF, Cosette Wheeler, Daron G Ferris, David Jenkins, Anne Schuind, Toufik Zahaf, Bruce Innis, Paulo Naud, Newton S De Carvalho, et al. Efficacy of a bivalent L1 virus-like particle vaccine in prevention of infection with human papillomavirus types 16 and 18 in young women: a randomised controlled trial. The Lancet. 364(9447):1757–1765. 2004.

4). Eaton L. Quantification of residual Escherichia coli DNA in recombinant biopharmaceutical proteins by hybridization analysis. J Pharm Biomed Anal. 7:633–638. 1989.
5). Europe Co. Recombinant DNA technology. European Pharmacopoeia Strasbourg: 18–26. 2001.
6). Food and Drug Administration. Points to consider in the production and testing of new drugs and biologicals produced by recombinant DNA technology. Office of Biologics Research and Review, Center for Drugs and Biologics, April 10. 1985.
7). Francoise B, Pierre C. Human papillomavirus vaccines. Seminars in Cancer Biology. 9:431–445. 1999.

8). Grachev V, Magrath D. WHO Requirements for the Use of Animal Cells as in vitro Substrates for the Production of Biologicals (Requirements for Biological Susbstances No. 50). Biologicals. 26(3):175–193. 1998.
9). Gregory CA, Rigg GP, Illidge CM, Matthews RC. Quantification of Escherichia coli Genomic DNA Contamination in Recombinant Protein Preparations by Polymerase Chain Reaction and Affinity-Based Collection. Analytical Biochemistry. 296(1):114–121. 2001.
10). Hagensee ME, Yaegashi N, Galloway DA. Self-assembly of human papillomavirus type 1 capsids by expression of the L1 protein alone or by coexpression of the L1 and L2 capsid proteins. J Virol. 67:315–322. 1993.

11). Harper DM, Franco EL, Wheeler C, Ferris DG, Jenkins D, Schuind A, Zahaf T, Innis B, Naud P, De Carvalho NS, Roteli-Martins CM, Teixeira J, Blatter MM, Korn AP, Quint W, Dubin G. Efficacy of a bivalent L1 virus-like particle vaccine in prevention of infection with human papillomavirus types 16 and 18 in young women: a randomised controlled trial. The Lancet. 364(9447):1757–1765. 2004.

12). Heino P, Dillner J, Schwartz S. Human Papillomavirus Type 16 Capsid Proteins Produced from Recombinant Semliki Forest Virus Assemble into Virus-like Particles. Virology. 214(2):349–359. 1995.

14). International Conference on Harmonisation. Viral safety evaluation of biotechnology products derived from cell lines of human or animal origin. ICH harmonised tripartite guideline. 1997.
15). Jeong HS, Shin JH, Park YN, Choi JY, Kim YL, Kim BG, Ryu SR, Baek SY, Lee SH, Park SN. Development of realtime RT-PCR for evaluation of JEV clearance during purification of HPV type 16 L1 virus-like particles. Biologicals. 31(3):223–229. 2003.

16). Kirnbauer R, Taub J, Greenstone H, Roden R, Durst M, Gissmann L, Lowy D, Schiller J. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J Virol. 67:6929–6936. 1993.

17). Koutsky LA AK, WHeeler CM, Brown DR, Barr E, Alvarez FB, Chiacchier ini LM, Jansen KU. A controlled trial of a human papillomavirus type 16 vaccine. N Engl J Med. 347(21):1703–1705. 2002; Nov 21.

18). Krause PR, Lewis J, Andrew M. Safety of Viral DNA in Biological Products. Biologicals. 26(4):317–320. 1998.

19). Lahijani R, Duhon M, Lusby E, Betita H, Marquet M. Quantitation of host cell DNA contaminate in pharmaceutical-grade plasmid DNA using competitive polymerase chain reaction and enzyme-linked immunosorbent assay. Hum Gene Ther. 9:1173–1180. 1998.

20). Lokteff M, Klinguer-Hamour C, Julien E, Picot D, Lannes L, Nguyen T, Bonnefoy JY, Beck A. Residual DNA Quantification in Clinical Batches of BBG2Na, a Recombinant Subunit Vaccine Against Human Respiratory Syncytial Virus. Biologicals. 29(2):123–132. 2001.

21). Pepin RA, Lucas DJ, Lang RB, Lee N, Liao MJ, Testa D. Detection of picogram amounts of nucleic acid by dot blot hybridization. Biotechniques. 8(6):628–632. 1990.
22). Perrin P, Morgeaux S. Inactivation of DNA by [beta]-propiolactone. Biologicals. 23(3):207–211. 1995.
24). Petricciani JC, Horaud FN. DNA, Dragones and Sanity. Biologicals. 23:233–238. 1995.
25). Pisani P, Parkin DM, Munoz N., Ferlay J. Cancer and Infection. Cancer Epidemiol Biomark Prev. 6:387–400. 1997.
26). Riggin A, Luu VT, Lobdell JK, Wind MK. A non-isotopic probe-hybridization assay for residual DNA in biopharmaceuticals. J Pharm Biomed Anal. 16(4):561–572. 1997.

27). Robertson JS, Heath AB. A Collaborative Study on DNA Quantitation in Biological Products. Biologicals. 23(3):199–205. 1995.

28). Roche Applied Science. DIG Application Manual for Nonradioactive In situ Hybridization. Application Manual. 3rd edition:. 9–13.
29). Sasagawa T, Pushko P, Steers G, Gschmeissner S, Hajibagheri M, Finch J, Crawford L, Tommasino M. Synthesis and assembly of virus-like particles of human papillomaviruses type 6 and type 16 in fission yeast Schizosaccharomyces pombe. Virology. 206:126–135. 1995.
30). Smith GR, Helf M, Nesbet C, Betita H, Meek J, Ferre F. Fast and accurate method for quantitating E. coli host-cell DNA contamination in plasmid DNA preparations. Biotechniques. 26:518–526. 1999.
31). Touze A, El Mehdaoui S, Sizaret PY, Mougin C, Munoz N, Coursaget P. The L1 major capsid protein of human papillomavirus type 16 variants affects yield of virus-like particles produced in an insect cell expression system. J Clin Microbiol. 36(7):2046–2051. 1998.

32). WHO Study Group. Acceptability of cell substrates for production of biologicals. WHO Technical Report Series No.747. 1987.
Go to : 
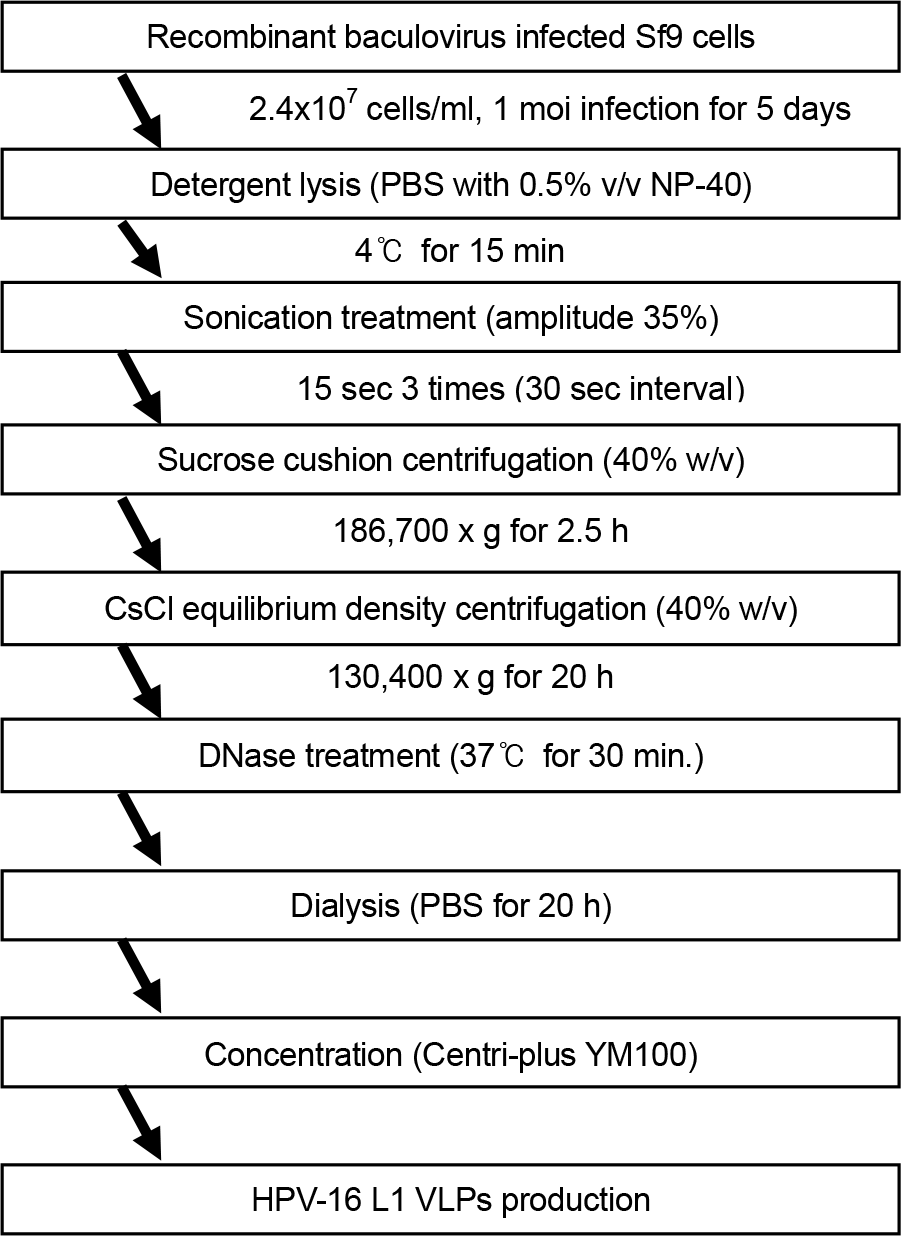 | Figure 1.Flowchart for the production of HPV-16 L1 VLPs. Each step was individually assessed for the ability to remove residual cellular DNA. |
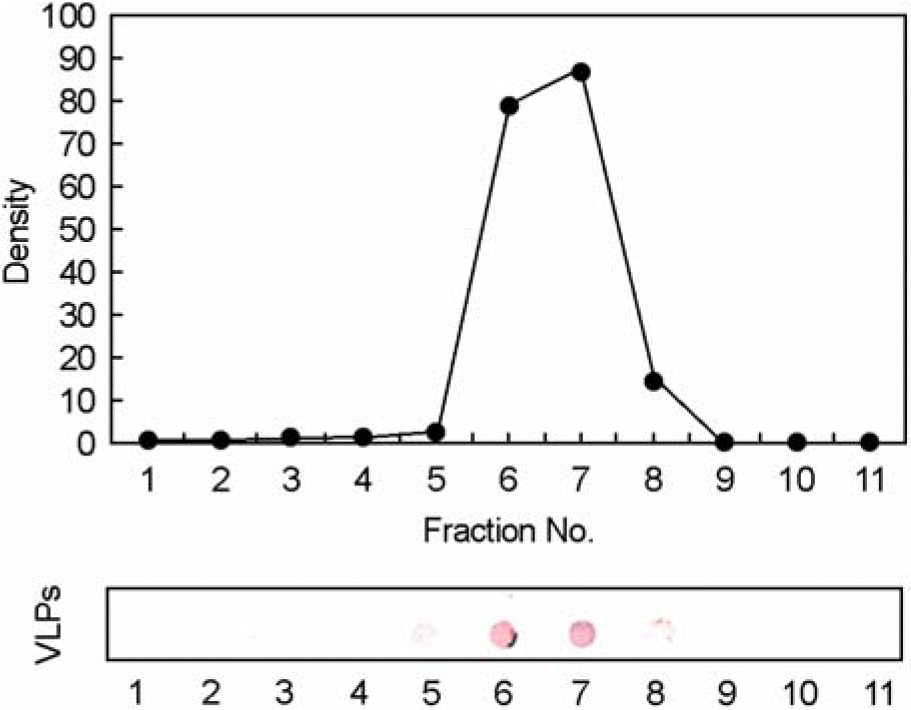 | Figure 2.Immunoblot analysis of HPV-16 type L1 VLPs in each fraction after CsCl equilibrium density centrifugation. The fractions containing VLPs were identified by immunoblot analysis using HPV-16 L1 monoclonal antibody, CamVir-1 and anti-mouse IgG alkaline phosphatase. |
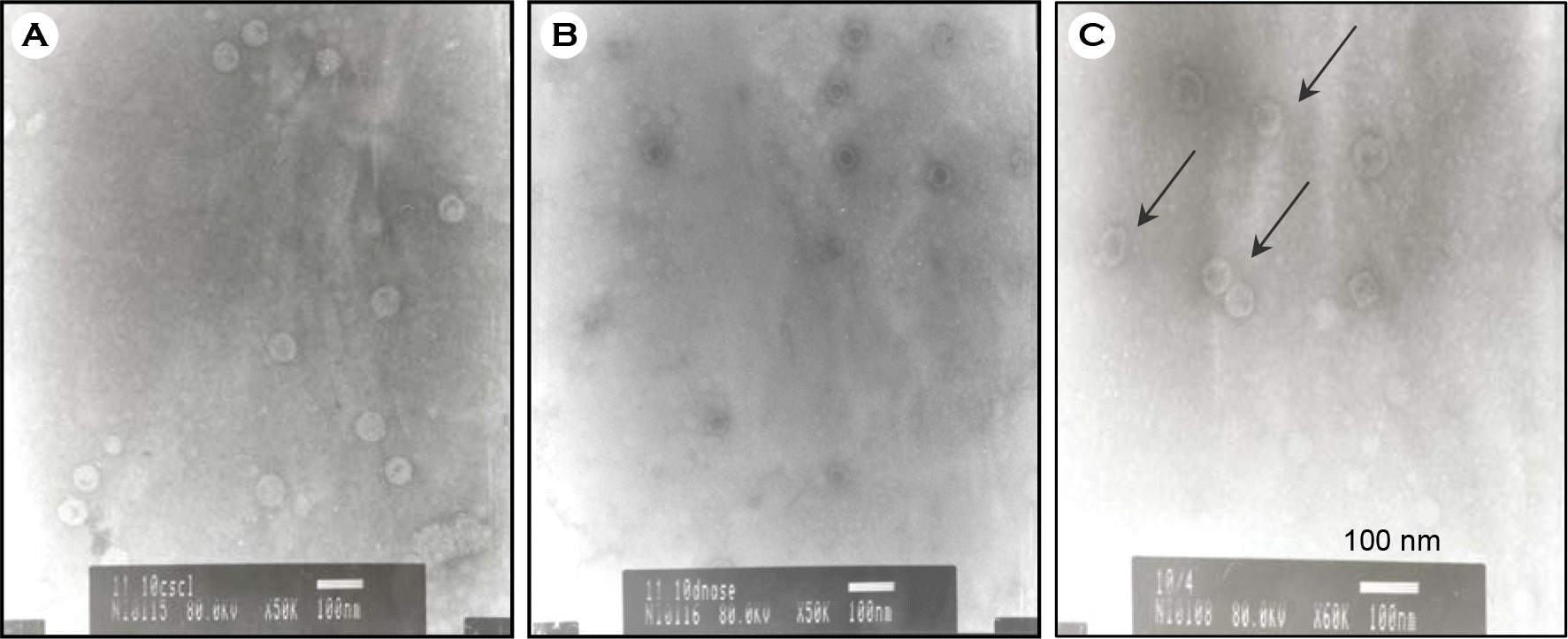 | Figure 3.Identification of HPV-16 L1 VLPs by electron micrograph. To confirm VLPs assembled, each preparation was applied to carbon-coated grids with an equal volume of VLPs suspension. After drying, the carbon-coated grids containing VLP were negatively stained with 2% uranyl acetate, and were observed at × 50,000 or × 60,000 electron microscope (JEOL.JP/JEM-1010, Japan). (A) CsCl ultracentrifuged sample; (B) DNase treated sample; and (C) final product. Arrows indicate examples of assembled HPV VLPs. Assembled VLPs were shown in all three samples (A, B, C). |
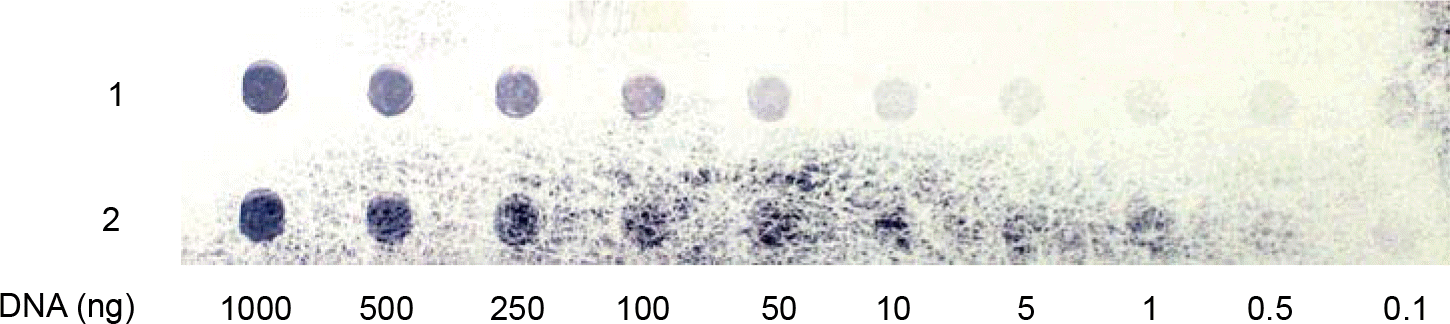 | Figure 4.Detection limit of residual cellular DNA. The limit of detection of the hybridization assay was evaluated at 0.1 ng with Random primed labeling kit. The figure shows 1000, 500, 100, 50, 10, 5, 1, 0.5, and 0.1 ng of targeted host cell derived-DNA (Sf9) spotted onto nylon membrane and assayed as described in materials and methods section. Detection limit of residual cellular DNA was 0.1 ng in this assay. |
 | Figure 5.Reproducibility and repeatability studies of hybridization assay. In order to validate the detection limit at 0.1 ng of random primed labeling DNA using the host-derived DNA, the reproducibility test was carried out on three different days. For the repeatability test, the DNA of 0.1 ng was blotted four times onto two separate membranes. |
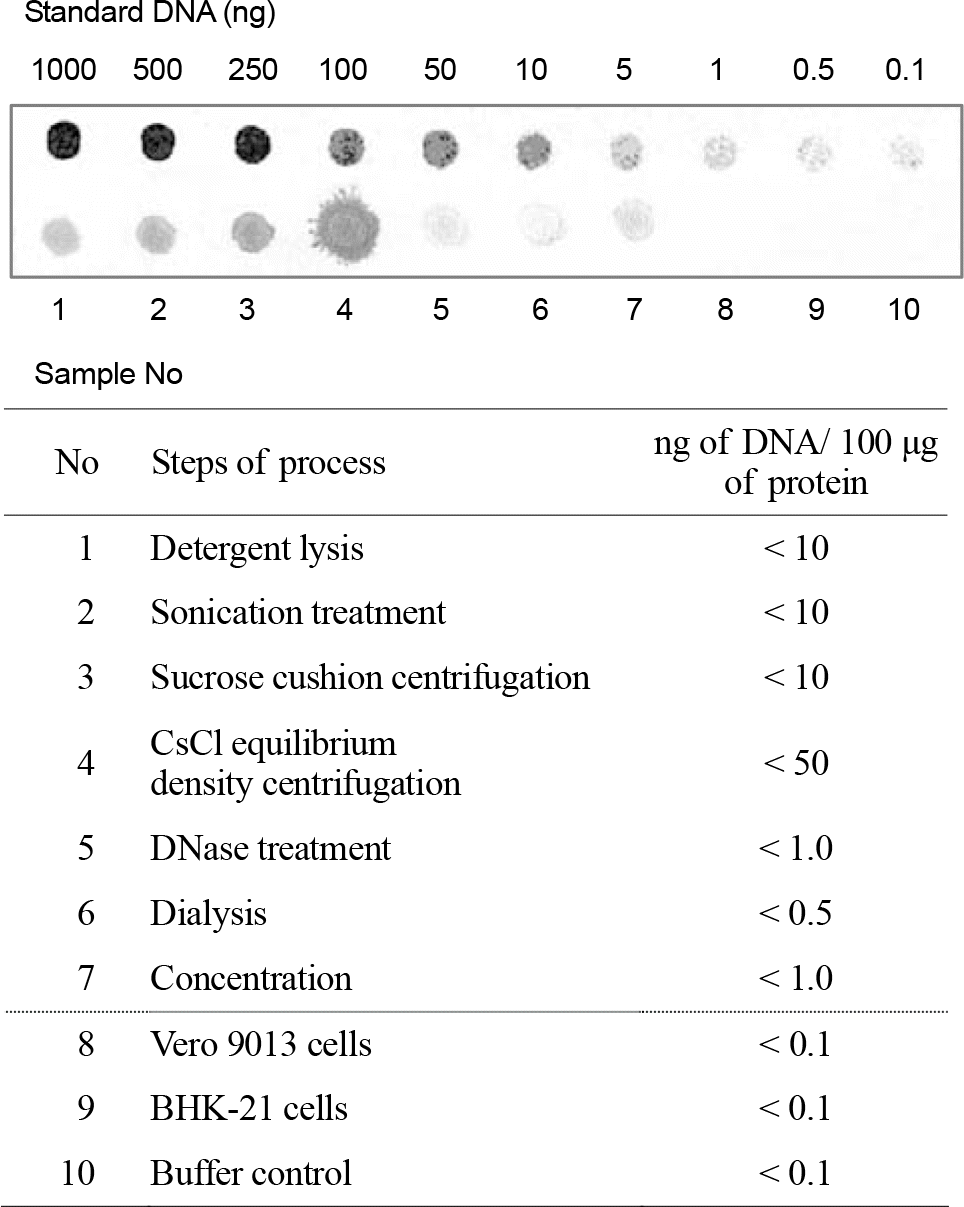 | Figure 6.Dot-blot hybridization assay for in process samples and final product. Compared to the standard DNA, the amount of residual cellular DNA in each process sample during purification process and in final product was determined (the amount of residual DNA in final product was 0.5 ng∼1 ng per 100 μg of protein). Vero 9013 cells and BHK-21 cells were used for controls of specificity test in the hybridization assay. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download