Abstract
G and P tying of group A porcine rotaviruses (PoRV) from field fecal samples were performed using reverse-transcriptase polymerization chain reaction (RT-PCR) and restriction fragment length polymorphism (RFLP) analysis. After amplifying full length VP7 and partial length VP4 genes, restriction endonucleases were used to digest and analyze the cutting pattern of the gene products. After analysis of digests with restriction endonucleases, seven and six RFLP types were observed for VP7 and VP4, respectively. The G typing analysis of 50 fecal samples revealed that 68% (34/50) were G4, which included G4-like (22/50); 22% (11/50) were G5; 6% (3/50) were G4 and G5 mixed types. The P typing analysis of the same fecal samples revealed that 36% (18/50) were P2B, 52% (26/50) were P9, 1 sample (2%) was a mixture of P2B and P9. Combinations of G and P types, the G4P2B and G4P9 types including G4-like accounted for 26% (13/50) and 32% (16/50), respectively. The G5P2B and G5P9 type also represented 4% (2/50) and 18% (9/50) of the samples. No G3 and G11 or other new P types were identified from the samples tested. Information on the G and P types and G/P combinations in the field fecal samples is useful for developing more effective PoRV vaccines and understanding the epidemiology of PoRV infections in the field.
References
1). Bohl EH, Theil KW, Saif LJ. Isolation and serotyping of porcine rotaviruses and antigenic comparison with other rotaviruses. J Clin Microbiol. 19:105–111. 1984.

2). Chang KO, Parwani AV, Saif LJ. The characterization of VP7 (G type) and VP4 (P type) genes of bovine group A rotaviruses from field samples using RT-PCR and RFLP analysis. Arch Virol. 141:1727–739. 1996.

3). Ciarlet M, Liprandi F. Serological and genomic characterization of two porcine rotaviruses with serotype G1 specificity. J Clin Microbiol. 32:269–272. 1994.

5). Estes MK, Palmer EL, Obijeski JF. Rotaviruses: a review. Curr Top Microbiol Immunol. 105:123–184. 1983.

6). Gomara MI, Wong C, Blome S, Desselberger U, Gray J. Rotavirus subgroup characterization by restriction endonuclease digestion of a cDNA fragment of the VP6 gene. J Virol Methods. 105:99–103. 2002.
7). Gouvea V, Santos N, Timenetsky Mdo C. VP4 typing of bovine and porcine group A rotaviruses by PCR. J Clin Microbiol. 32:1333–1337. 1994.

8). Gouvea V, Santos N, Timenetsky MdoC. Identification of bovine and porcine rotavirus G types by PCR. J Clin Microbiol. 32:1338–1340. 1994.

9). Hoshino Y, Sereno MM, Midthun K, Flores J, Kapikian AZ, Chanock RM. Independent segregation of two antigenic specificities (VP3 and VP7) involved in neutralization of rotavirus infectivity. Proc Natl Acad Sci USA. 82:8701–8704. 1985.

10). Hoshino Y, Wyatt RG, Greenberg HB, Flores J, Kapikian AZ. Serotypic similarity and diversity of rotaviruses of mammalian and avian origin as studied by plaque-reduction neutralization. J Infect Dis. 149:694–702. 1984.
11). Kapikian AZ, Hoshino Y, Chanock RM. Rotaviruses. Knipe DM, Howley PM, editors. Fields Virology. 4th ed.pp.p. 1787–1833. Lippincott Williams & Wilkins;Philadelphia: 2001.
12). Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685. 1970.

13). Li B, Gorziglia M. VP4 serotype of the Gottfried strain of porcine rotavirus. J Clin Microbiol. 31:3075–3077. 1993.

14). Lucchelli A, Kang SY, Jayasekera MK, Parwani AV, Zeman DH, Saif LJ. A survey of G6 and G10 serotypes of group A bovine rotaviruses from diarrheic beef and dairy calves using monoclonal antibodies in ELISA. J Vet Diagn Invest. 6:175–181. 1994.

15). Nagesha HS, Holmes IH. New porcine rotavirus serotype antigenically related to human rotavirus serotype 3. J Clin Microbiol. 26:171–174. 1988.

16). Nishikawa K, Fukuhara N, Liprandi F, Green K, Kapikian AZ, Chanock RM, Gorziglia M. VP4 protein of porcine rotavirus strain OSU expressed by a baculovirus recombinant induces neutralizing antibodies. Virology. 173:631–637. 1989.

17). Offit PA, Blavat G. Identification of the two rotavirus genes determining neutralization specificities. J Virol. 57:376–378. 1986.

18). Parwani AV, Hussein HA, Rosen BI, Lucchelli A, Navarro L, Saif LJ. Characterization of field strains of group A bovine rotaviruses using polymerase chain reaction generated G and P type specific cDNA probes. J Clin Microbiol. 31:2010–2015. 1993.
19). Rosen BI, Parwani AV, Gorziglia M, Larralde G, Saif LJ. Characterization of full-length and polymerase chain reaction-derived partial-length Gottfried and OSU gene 4 probes for serotypic differentiation of porcine rotaviruses. J Clin Microbiol. 30:2644–2652. 1992.

20). Rosen BI, Saif LJ, Jackwood DJ, Gorziglia M. Serotypic differentiation of group A rotaviruses with porcine rotavirus gene 9 probes. J Clin Microbiol. 28:2526–2533. 1990.

21). Ruiz AM, Lopez IV, Lopez S, Espejo RT, Arias CF. Molecular and antigenic characterization of porcine rotavirus YM, a possible new rotavirus serotype. J Virol. 62:4331–4336. 1988.

22). Saif LJ, Rosen BI, Kang SY, Miller KL. Cell culture propagation of rotaviruses. J Tissue Culture Methods. 11:147–156. 1988.

23). Sereno MM, Gorziglia MI. The outer capsid protein VP4 of murine rotavirus strain Eb represents a tentative new P type. Virology. 199:500–504. 1994.

24). Song DS, Yang JS, Oh JS, Han JH, Park BK. Differentiation of a Vero cell adapted porcine epidemic diarrhea virus from Korean field strains by restriction fragment length polymorphism analysis of ORF 3. Vaccine. 21:1833–1842. 2003.

25). Taniguchi K, Urasawa T, Kobayashi N, Gorziglia M, Urasawa S. Nucleotide sequence of VP4 and VP7 genes of human rotaviruses with subgroup I specificity and long RNA pattern: implication of new G serotype specificity. J Virol. 64:5640–5644. 1990.
Figure 1.
Restriction endonuclease cleavage patterns of reference PoRV VP7 (A) and VP4 (B) genes.
M: Marker, E: EcoRI, B: BamHI, H: HindIII, N: NlaIV, V: VspI, R: EcoRV
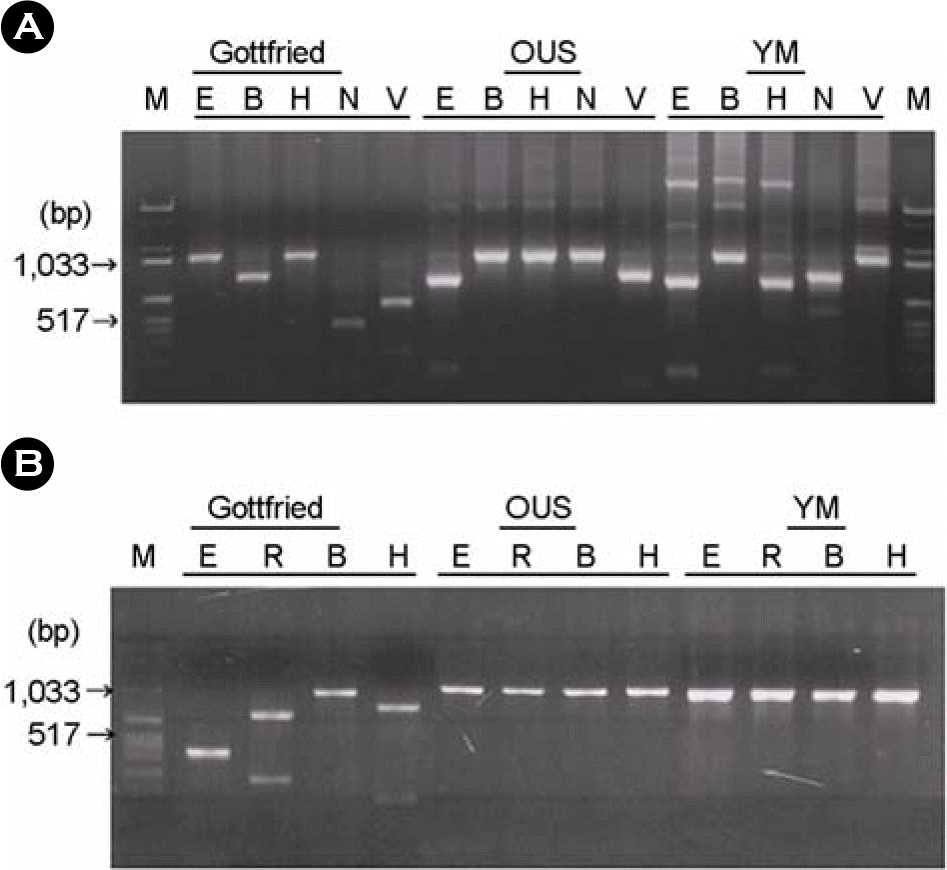
Figure 2.
Restriction endonuclease cleavage patterns of PoRV VP7 genes from fecal samples.
M: Marker, E: EcoRI, B: BamHI, H: HindIII, N: NlaIV, V: VspI I, IIa, IIb, IIIa, IIIb, IIIc and IV represent VP7 RFLP type.
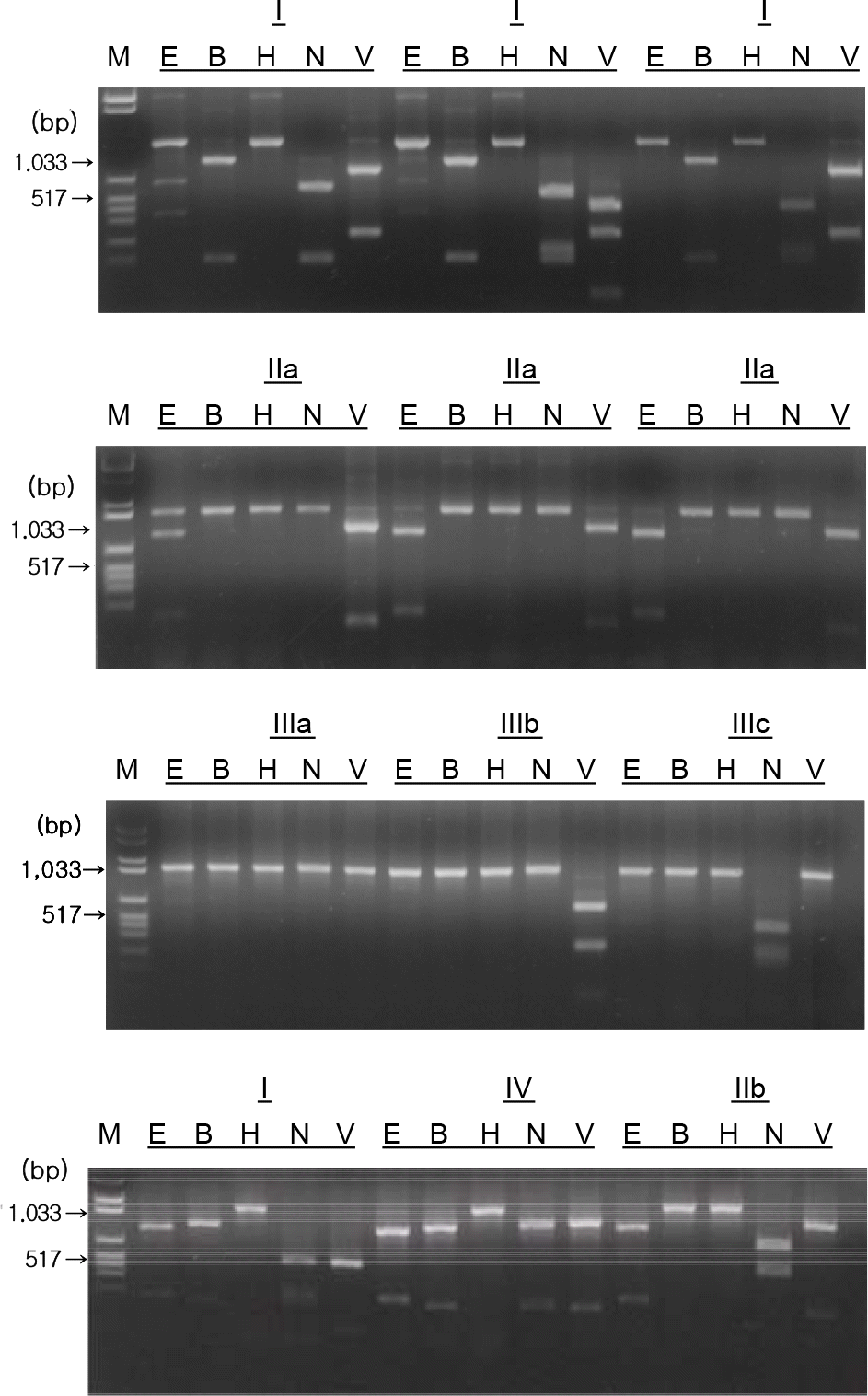
Figure 3.
Restriction endonuclease cleavage patterns of PoRV VP4 genes from fecal samples.
M: Marker, E: EcoRI, R: EcoRV, BamHI, H: HindIII 1, 2, 3, 4, 5, and 6 represent VP4 RFLP type.
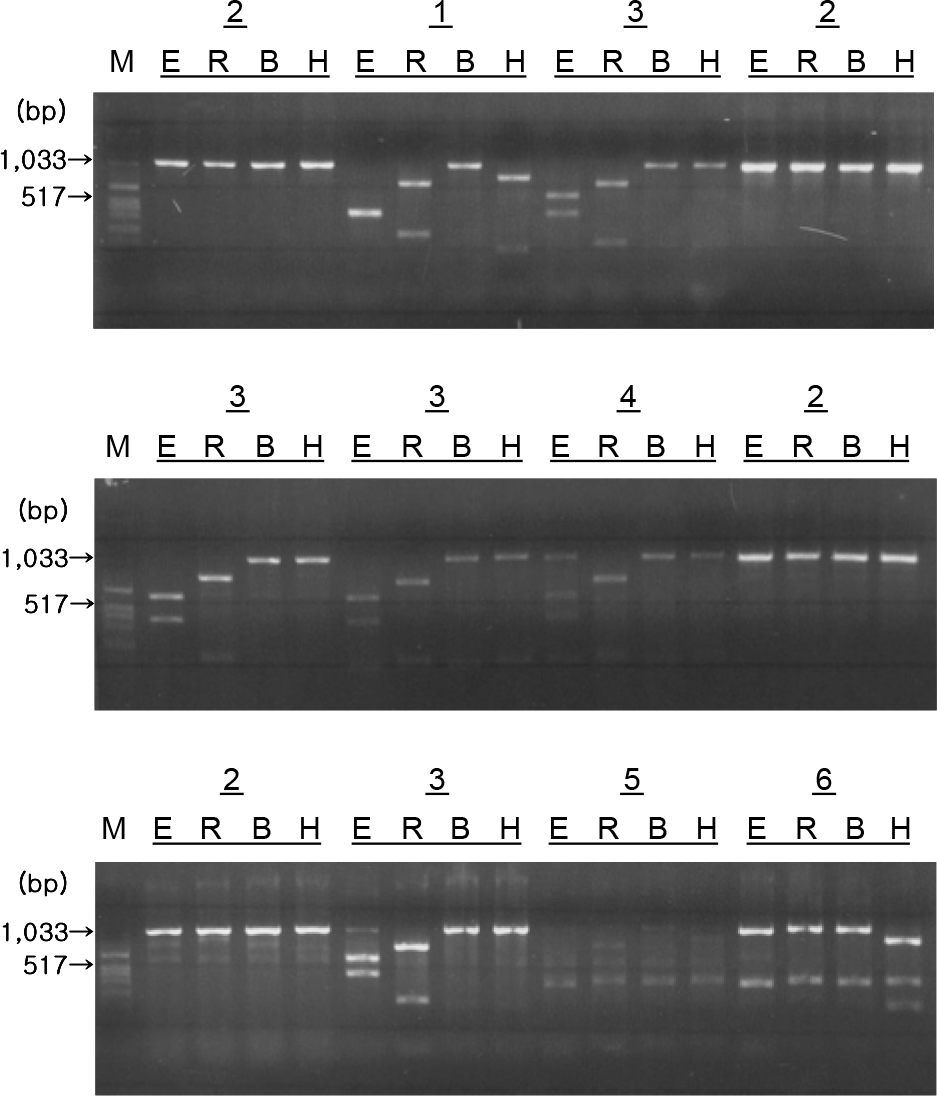
Table 1.
Restriction fragment length polymorphism analysis of the RT-PCR product of the VP7 gene from PoRV field samples
Table 2.
Restriction fragment length polymorphism analysis of the RT-PCR product of the VP4 gene from PoRV field samples
Table 3.
Comparison of the VP7 nucleotide sequences of the three PoRV field isolates with those of reference PoRV
| Isolate | Similarity (%) | |||||
|---|---|---|---|---|---|---|
| A | B | C | Gottfried | OSU | YM | |
| A (IIIb) | ▪ | 82.6 | 84.3 | 84.6 | 71.7 | 71.6 |
| B (IIIc) | ▪ | 82.0 | 83.6 | 72.5 | 71.9 | |
| C (IV) | ▪ | 94.2 | 73.5 | 71.1 | ||
| Gottfried | ▪ | 76.0 | 74.9 | |||
| OSU | ▪ | 72.9 | ||||
| YM | ▪ | |||||
Table 4.
G and P type combinations of PoRV field samples analyzed by RT-PCR and RFLP




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download