Abstract
Acinetobacter species are non-fermentative Gram-negative coccobacilli and they have emerged as important nosocomial pathogens which are associated with the significant multidrug resistance in recent years. Carbapenem-resistant A. baumannii (CRAB) and pandrug-resistant A. baumannii (PDRAB) were reported in 1991 and 1998, respectively. Fifty-eight isolates of Acinetobacter species recovered from a university hospital between August 2004 and March 2005 were investigated for the existence of CRAB, PDRAB, extended-spectrum β-lactamase (ESBL)-producing Acinetobacter and examined for their phenotypic and genotypic characteristics. Genomospecies of Acinetobacter species were determined by amplified rDNA restriction analysis (ARDRA) and antimicrobial susceptibility test was performed with 13 kinds of antimicrobial agents. Metallo-β-lactamase (MBL) producers were screened by modified hodge test and confirmed by imipenem-EDTA disk synergy test. Detection of blaIMP-1, blaVIM-2, blaTEM, and blaPER-1 was performed by PCR. Genomic DNAs were analyzed by pulsed-field gel electrophoresis (PFGE). Among 58 isolates of Acinteobacter species, 40 isolates were identified as genospecies 2 (A. baumannii), 9 were 13TU, 5 were A. phenon 6/ct, and 4 were Acinetobacter genospecies 3 by ARDRA. Thirteen isolates were confirmed as MBL-producers and blaIMP-1 and blaVIM-2 were carried by 5 and 8 isolates of them, respectively. MBL-producers were mostly 13TU, A. phenon 6/ct 13TU, and Acinetobacter genospecies 3 and they were susceptible to ciprofloxacin and ampicillin-sulbactam. BlaPER-1 was carried by thirteen isolates and 12 isolates of them were PDRAB showing resistance to all antimicrobial agents tested, including ceftazidime, cefepime, aztreonam, ciprofloxacin, amikacin, gentamicin, ampicillin-sulbactam, and imipenem. In conclusion, most MBL-producers belonged to 13TU, A. phenon 6/ct 13TU, and Acinetobacter genospecies 3 which were susceptible to ciprofloxacin and ampicillin-sulbactam, whereas 12 of 13 PER-1-producers were PDRAB originated from the same clone.
References
1). Aubert G, Guichard D, Vedel G. In vitro activity of cephalosporins alone and combinated with different antibiotic resistance profiles. J Antimicrob Chemother. 37:155–160. 1996.
2). Bergogne-Bérézin E, Joly-Guillou M, Vieu JF. Epidemiology of nosocomial infection due to Acinetobacter calcoaceticus. J Hosp Infect. 10:105–113. 1987.
3). Bergogne-Bérézin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 9:148–165. 1996.
4). Bouvet PJ, Grimont PA. Taxonomy of the Genus Acinetobacter with the Recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., Acinetobacter junii sp. nov., and Emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int J Bacteriol. 36:228–240. 1986.
5). Bouvet PJ, Jeanjean S. Delineation of new proteolytic genomic species in the genus Acinetobacter. Res Microbiol. 140:291–299. 1989.
6). Bradford PA. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology therapy, and detection of this important resistance threat. Clin Microbiol Rev. 14:933–951. 2001.
7). Carr EL, Kampfer P, Patel BK, Gurtler V, Seviour RJ. Seven novel species of Acinetobacter isolated from activated sludge. Int J Syst Evol Microbiol. 53:953–963. 2003.
8). Corbella X, Montero A, Pujol M, Domiguez MA, Ayats J, Argerich MJ, Garrigosa F, Ariza J, Gudiol F. Emergence and rapid spread of carbapenem resistance during a large and sustained hospital outbreak of multiresistant Acinetobacter baumannii. J Clin Microbiol. 38:4086–4095. 2000.
9). Gautom RK. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J Clin Microbiol. 35:2977–2980. 1997.
10). Gerner-Smidt P, Tjernberg I. Acinetobacter in Denmark. II. Molecular studies of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex. Acta Pathol Microbiol Immunol Scand. 101:826–832. 1993.
11). Hsueh PR, Teng LJ, Chen CY, Chen WH, Yu CJ, Ho SW, Luh KT. Pandrug-resistant Acinetobacter baumannii causing nosocomial infections in a university hospital, Taiwan. Emerg Infect Dis. 8:827–832. 2002.
12). Iyobe S, Kusadokoro H, Ozaki J, Matsumura N, Minami S, Haruta S, Sawai T, O'Hara K. Amino acid substitution in a variant of IMP-1 metallo-β-lactamase. Antimicrob Agents Chemother. 44:2023–2027. 2000.
13). Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini GM. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 43:1584–1590. 1999.
14). Lee K, Chong Y, Shin HB, Kim YA Yong D, Yum JH. Modified Hodge and EDTA-disk synergy tests to screen metallo-β-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 7:88–91. 2001.
15). Lee K, Chong Y, Shin HB, Yong D. Rapid increase of imipenem-hydrolyzing Pseudomonas aeruginosa in a Korean hospital. Abstr. E-85, 38th ICAAC. 1998.
16). Lee K, Ha GY, Shin BM, Kim JJ, Kang JO, Jang SJ, Yong D, Chong Y. Korean Nationwide Surveillance of Antimicrobial Resistance Group. Metallo-β-lactamase-producing Gram-negative bacilli in Korean Nationwide Surveillance of Antimicrobial Resistance group hospitals in 2003. Diag Microb Inf Dis. 50:51–58. 2004.
17). Lee K, Jang SJ, Lee HJ, Ryoo N, Kim M, Hong SG. Korean Nationwide Surveillance of Antimicrobial Resistance Group. Increasing prevalence of vancomycin-resistant enterococci, expanded cephalosporin-resistant Klebiella pneumoniae, and multidrug-resistant Pseudomonas aeruginosa and Acinetobacters in Korea: 2001 KONSAR study. J Korean Med Sci. 19:8–14. 2004.
18). Lee K, Lee HS, Jang SJ, Park AJ, Lee MH, Song WK, Chong Y. Korean Nationwide Surveillance of Antimicrobial Resistance Group. Antimicrobial Resistance Surveillance of Bacteria in 1999 in Korea with a Special Reference to Resistance of Enterococci to Vancomycin and Gram-Negative Bacilli to Third Generation Cephalosporin, Imipenem, and Fluoroquinolone. J Korean Med Sci. 16:262–270. 2001.

19). Lee K, Lim JB, Yum JH, Yong D, Chong Y, Kim JM, Livermore DM. blaVIM-2 cassette containing novel integrons in metallo-β-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob Agents Chemother. 46:1053–1058. 2002.
20). Lee K, Yong D, Kim DS, Kim SK, Pai CH, Oh HB, Yum JH, Chong Y, Cho DT. Antimicrobial resistance of clinical isolates of bacteria isolated from January to March in 2005 at Severance Hospital, Seoul. Antimicrobial Resistance. Newsletter. 13(2):1. 2005.
21). Luzzaro F, Mantengoli E, Perilli M, Lombardi G, Orlandi V, Orsatti A, Amicosante G, Rossolini GM, Toniolo A. Dynamics of a nosocomial outbreak of multidrug-resistant Pseudomonas aeruginosa producing the PER-1 extended-spectrum β-lactamase. J Clin Microbiol. 39:1865–1870. 2001.
22). National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 6th ed.Approved standard M7-A6;Wayne, PA: 2003.
23). Nemec A, De Baere T, Tjernberg I, Vaneechoutte M, van der Reijden TJK, Dijchoorn L. Acinetobacter ursingii sp. nov. and Acinetobacter schindleri sp. nov., isolated from human clinical specimens. Int J Syst Evol Microbiol. 51:1891–1899. 2001.
24). Nordmann P, Ronco E, Naas T, Duport C, Michel-Briand Y, Labia R. Characterization of a novel extended-spectrumlactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 37:962–969. 1993.
25). Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, Yoshimura F, Kato N. Molecular characterization of an enterobacterial metallo-β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob Agents Chemother. 38:71–78. 1994.
26). Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo J-D, Nordmann P. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-lactamase and its plasmid- and integronborne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob Agents Chemother. 44:891–897. 2000.
27). Schreckenberger PC, Von Graevenitz A. Acinetobacter, Achromobacter, Alcaligenes, Moraxella, Methylobacterium and other non-fermentative Gram-negative rods. Manual of clinical microbiology. 7th edn. Washington, DC: American Society for Microbiology; p.539–560. 1990.
28). Simor AE, Lee M, Vearncombe M, Jones-Paul L, Barry C, Gomez M, Fish JS, Cartotto RC, Palmer R, Louie M. An outbreak due to multiresistant Acinetobacter baumannii in a burn unit: risk factors for acquisition and management. Infect Control Hosp Epidemiol. 23:261–267. 2002.
29). Tjernberg I. Antimicrobial susceptibility of Acinetobacter strains identified by DNA-DNA hybridization. APMIS. 98:320–326. 1990.
30). Tjerberg I, Ursing J. Clinical strains of Acinetobacter classified by DNA-DNA hybridization. Acta Pathol Microbiol Immunol Scand. 97:595–605. 1989.
31). Vahaboglu H, Ozturk R, Aygun G, Coskunkan F, Yaman F, Kaygusuz A, Leblebicioglu H, Balik I, Aydin K, Otkun M. Widespread detection of PER-1-type extended-spectrum β-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multi-center study. Antimicrob Agents Chemother. 41:2265–2269. 1997.
32). Vaneechoutte M, Dijishoorn L, Tjernberg I, Elaichouni A, de Vos P, Claeys G, Verschraegen G. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J Clin Microbial. 33:11–15. 1995.
33). Vaneechoutte M, Tjerberg I, Baldi F, Pepi M, Fani R, Sullivan ER, van der Toorn J, Dijkshoorm L. Oil-degrading Acinetobacter strain RAG-1 and strains described as ‘Acinetobacter venetianus sp. nov.’ belong to the same genomic species. Res Microbiol. 150:69–73. 1999.
34). Yong D, Shin JH, Kim S, Lim Y, Yum JH, Lee K, Chong Y, Bauernfeind A. High Prevalence of PER-1 extended-spectrum β-lactamase-producing Acinetobacter spp. in Korea. Antimicrob Agents Chemother. 5:1749–1751. 2003.
Figure 1.
Restriction patterns obtained after restriction digestion with CfoI, AluI, MboI, RsaI, and MspI for amplified 16S rDNA of different Acinetobacter species. Numbers below each lane correspond to arbitrarily assigned ARDRA pattern numbers for each enzyme (http://users.ugent.be/∼mvaneech/ARDRA/Acinetobacter.html).
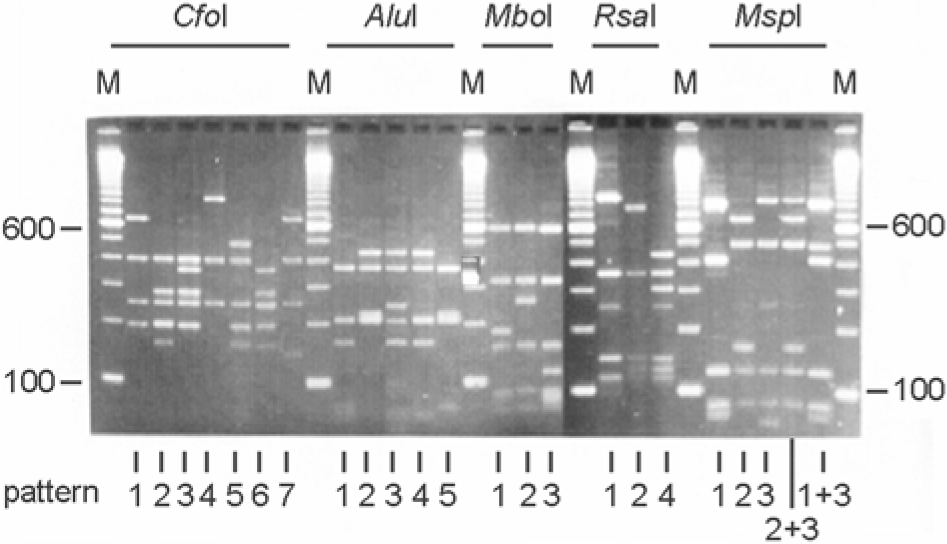
Figure 2.
Dendrogram generated by Gel Compar II software showing the relatedness of Acinetobacter baumannii determined by PFGE.
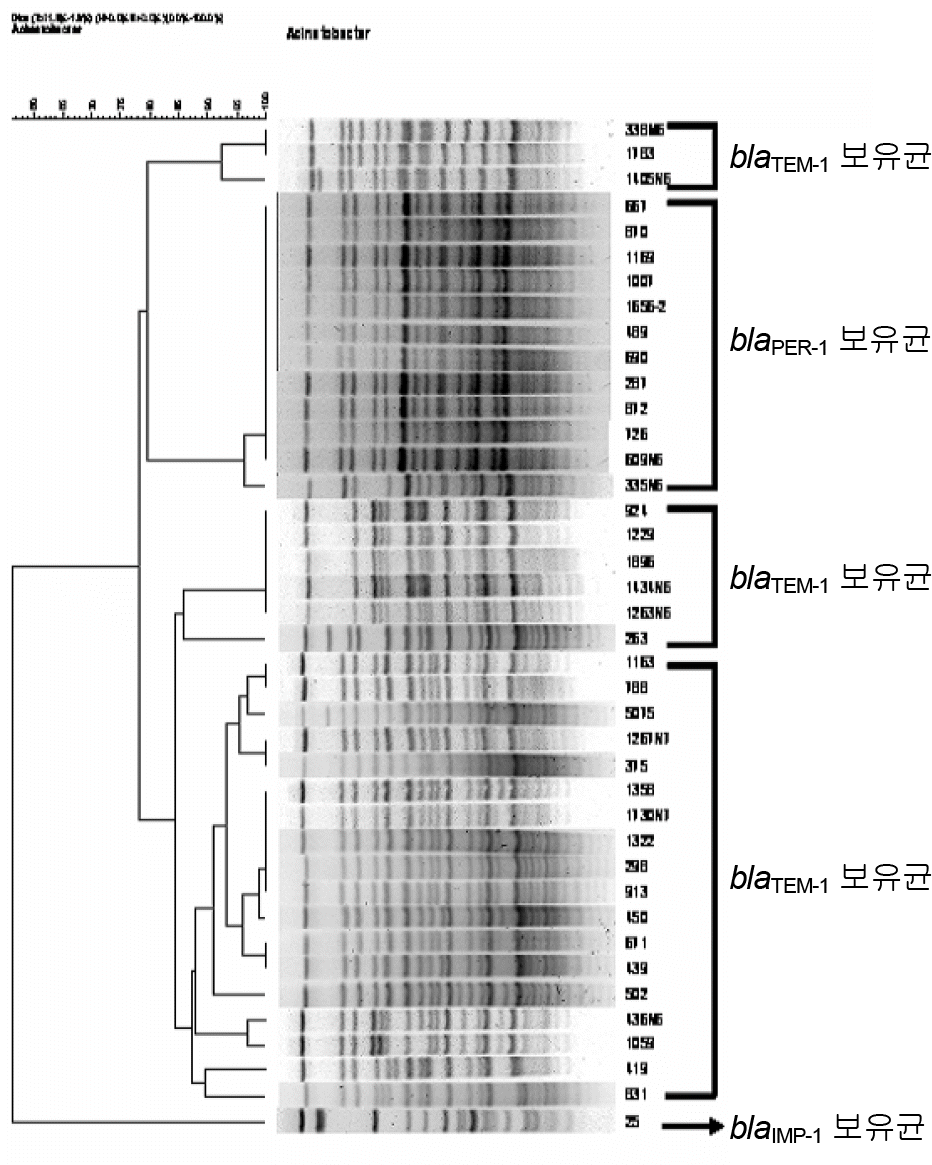
Table 1.
Oligonucletide primers used for detection of β-lactamase genes
| Primers | Tma (°C) | Nucleotide sequence | References | Expected amplicon size (bp) |
|---|---|---|---|---|
| TEM-sence | 5′-ATA AAA TTC TTG AAG ACG AAA-3′ | |||
| TEM-antisence | 50 | 5′-GAC AGT TAC CAA TGC TTA ATC-3′ | AB103506 | 1080 |
| SHV-sence | 5′-TGG TTA TGC GTT ATA TTC GCC-3′ | |||
| SHV-antisence | 55 | 5′-GGT TAG CGT TGC CAG TGC T-3′ | AY223863 | 869 |
| CTX-M-sence | 5′-CGC TTT ATG CGC AGA CGA-3′ | |||
| CTX-M-antisence | 55 | 5′-GAT TCT CGC CGC TGA AGC-3′ | AY847147 | 785 |
| OXA-1-sence' | 5′-AGC CGT TAA AAT TAA GCC C-3′ | |||
| OXA-1-antisence | 55 | 5′-CTT GAT TGA AGG GTT GGG CG-3′ | AV162283 | 908 |
| PER-1-sence | 5′-ATG AAT GTC ATT ATA AAA GC-3′ | |||
| PER-1-antisence | 50 | 5′-AAT TTG GGC TTA GGG CAA GAA A-3′ | Z21957 | 925 |
| VIM-2-sence | 5′-ATT GGT CTA TTT GAC CGC GTC-3′ | |||
| VIM-2-antisence | 54 | 5′-TGC TAC TCA ACG ACT GAC CG-3′ | DQ153217 | 780 |
| IMP-1-sence | 5′-CTA CCG CAG CAG ACT CTT TGC-3′ | |||
| IMP-1-antisence | 55 | 5′-GAA CAA CCA GTT TTG CCT TAC C-3′ | AB162949 | 591 |
Table 2.
ARDRA profiles of Acinetobacter species
| Genomic species | Pattern with enzymea | No. of isolates | Reference strain | ||||
|---|---|---|---|---|---|---|---|
| CfoI | AluI | MboI | RsaI | MspI | |||
| 2 (A. baumannii) | 1 | 1 | 1 | 2 | 3 | 39 | ATCC 17904 |
| 1 | 1 | 1 | 2 | 1 | 1 | ATCC 19606 | |
| 3 | 2 | 1 | 3 | 1 | 3 | 5 | ATCC 19004 |
| 13TU | 2 | 1 | 1 | 1 | 1 | 3 | ATCC 17903 |
| 2 | 1 | 1 | 1 | 3 | 4 | 100 | |
| 2 | 1 | 1 | 1 | 1+3 | 2 | RUH 2624 | |
| A. phenon 6/ct 3TU | 3 | 1 | 3 | 1 | 3 | 4 | MGH 99896, 5804 |
Table 3.
Antimicrobial resistance for clinical isolates of Acinetobacter species
| Antibiotics | No. of resistant strains | Total (n= 58) | |||
|---|---|---|---|---|---|
| A. baumannii (n=40) | 13TU (n=9) | A. phenon 6/ct 13TU (n=5) | Acinetobacter genospecies 3 (n=4) | ||
| piperacillin | 38 | 4 | 4 | 3 | 49 (84.5%) |
| ticarcillin | 38 | 4 | 2 | 2 | 46 (79.3%) |
| amikacin | 39 | 7 | 2 | 3 | 51 (87.9%) |
| gentamicin | 40 | 8 | 5 | 3 | 56 (96.6%) |
| tobramycin | 40 | 7 | 5 | 3 | 55 (94.8%) |
| ciprofloxacin | 39 | 1 | 0 | 0 | 40 (69.0%) |
| tazobactam | 35 | 2 | 1 | 2 | 40 (69.0%) |
| ceftazidime | 39 | 7 | 0 | 3 | 49 (84.5%) |
| cefepime | 35 | 3 | 0 | 3 | 41 (70.7%) |
| aztreonam | 36 | 5 | 0 | 2 | 43 (74.1%) |
| imipenem | 13 | 5 | 5 | 2 | 25 (43.0%) |
| meropenem | 14 | 4 | 1 | 2 | 21 (36.2%) |
| amp/sula | 38 | 2 | 0 | 0 | 40 (69.0%) |
Table 4.
Antimicrobial susceptibility to imipenem, modified Hodge test, and detection of metallo-β-lactamases and β-lactamases among clinical isolates of Acinetobacter spp.
Table 5.
Antimicrobial susceptibilities of β-lactamase or metallo-β-lactamase producing Acinetobacter species
| Antibiotics | MICa (μg/ml) for isolates | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| blaPER-1 positive (n=13) | blaTEM-1 positive (n=27) | blaVIM-2 positive (n=8) | blaIMP-1 positive (n=5) | |||||||||
| Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | |
| piperacillin | 32∼512 | 256 | 512 | 512∼>512 | >512 | >512 | 32∼256 | 64 | 256 | 16∼256 | 64 | 256 |
| amikacin | 64∼128 | 64 | 64 | 4∼>256 | >256 | >256 | 32∼128 | 64 | 128 | 64∼256 | 256 | 256 |
| gentamicin | >64 | >64 | >64 | >64 | >64 | >64 | 32∼>64 | >64 | >64 | 8∼64 | 32 | 64 |
| tobramycin | >64 | >64 | >64 | 64∼>64 | >64 | >64 | 32∼>64 | 64 | >64 | 64∼>64 | >64 | >64 |
| ciprofloxacin | 0.5∼>16 | >16 | >16 | 16∼>16 | >16 | >16 | <0.25∼0.5 | 0.25 | 0.5 | <0.25∼2 | 0.25 | 2 |
| amp-sulb | 16∼128 | 32 | 128 | 16∼>128 | 64 | 128 | 4∼32 | 8 | 32 | 1∼16 | 2 | 16 |
| ceftazidime | >128 | >128 | >128 | 16∼>128 | >128 | >128 | 16∼64 | 32 | 64 | 128∼>128 | >128 | >128 |
| cefepime | >128 | >128 | >128 | 16∼128 | 64 | 128 | 4∼16 | 16 | 16 | 16∼128 | 64 | 128 |
| aztreonam | >128 | >128 | >128 | 16∼128 | 64 | 128 | 32∼64 | 32 | 64 | 16∼32 | 16 | 32 |
| imipenem | 1∼16 | 16 | 16 | 2∼8 | 2 | 8 | 16∼64 | 16 | 64 | 16∼64 | 32 | 64 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download