Abstract
Porcine circovirus type 2 (PCV-2) has been nowadays recognized as a major agent causing postweaning multisystemic wasting syndrome (PMWS) in pigs worldwide. PMWS most commonly affects the weaned piglets, being of increasing importance to the pig industry in Korea. Seven commercial farms affected with PMWS and 2 farms free from PMWS, located in the southern part of Gyeonggi province, were selected for this study. The peripheral mononuclear cells were tested for the presence of ORF2 gene by PCR, and 54 (68.4%) of 79 samples were positive. All of 9 herds tested included the positive cases. The positive rates by herds were 50 to 100% in the PMWS-affected herds and 40 to 62.5% in the PMWS-free herd. The nucleotide and amino acid sequences of ORF2 gene of 6 strains were characterized. Homologies among 6 strains revealed 92.1 to 100% in the nucleotide and 92.3 to 100% in the amino acid. The overall ranges of homologies for 25 strains comprised 2 Korean and 23 foreign strains were 91.1 to 100% in the nucleotide and 89.7 to 100% in the amino acid. Three regions of greater heterogeneity were found in immunorelevant epitopes of the capsid protein, and the sequences between 57 to 80 aa revealed higher mutation than other areas. In the phylogenetic tree analysis, KOR 71 strain was clustered together with Korean strains previously isolated in Korea. The remaining 5 strains were closely clustered with other European and Asian strains. The results will be valuable for improving our understanding of the molecular epidemiology of PCV-2 in Korea and development of preventive measures for PMWS.
References
2). Allan GM, Meehan B, Todd D, Kennedy S, McNeilly F, Ellis J, Clark EG, Harding J, Espuna E, Botner A, Charreyre C. Novel porcine circoviruses from pigs with wasting disease syndromes. Vet Rec. 142:467–468. 1998.
3). de Boisseson C, Beven V, Bigarre L, Thiery R, Rose N, Eveno E, Madec F, Jestin A. Molecular characterization of porcine circovirus type 2 isolates from post-weaning multisystemic wasting syndrome-affected pigs. J Gen Virol. 85:293–304. 2004.
4). Chae C. Postweaning multisystemic wasting syndrome: a review of etiology, diagnosis and pathology. Vet J. 168:41–49. 2004.
5). Choi C, Chae C. In-situ hybridization for the detection of porcine circovirus 2-infected pigs with postweaning multisystemic wasting syndrome as shown by in-situ hybridization. J Com Patho. 121:265–270. 1999.
6). Clark EG. Post-weaning multisystemic wasting syndromes. Proc Am Ass Swine Pract. 10:499–501. 1997.
7). Dominique M, Philippe B, Catherine T, Claire A, Pierre LC, Roland C, Francois M, Emmanuel A, Andre J. Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J Gen Virol. 81:1815–1824. 2000.
8). Ellis J, Hassard L, clark E, Harding J, Allan G, Willson P, Storkappe J, Martine K, McNeilly F, Meehan B, Todd D, Haines D. Isolation of circovirus from lesion of pigs with postweaning multisystemic wasting syndrome. Can Vet J. 39:44–51. 1998.
9). Fenaux M, Halbur P, Gill M, Toth TE, Meng XJ. Genetic characterization of type 2 porcine circovirus (PCV-2) from pigs with multisystemic wasting syndrome in different geographic regions of North America and length polymorphism assay to detect and differentiate between infections with PCV-1 and PCV-2. J Clin Microbiol. 38:2494–2503. 2000.
10). Garkavenko O, Elliott RB, Croxson MC. Identification of pig circovirus Type 2 in New Zealand pigs. Transplant Proc. 37:506–509. 2005.

11). Hamel AL, Lin LL, Nayar GPS. Nucleotide sequence of porcine circovirus associated with postweaning multi-systemic wasting syndrome in pigs. J Virol. 72:5262–5267. 1998.

12). Hamel AL, Lin LL, Sachive C, Grudeski E, Nayar GPS. PCR detection and characterization of type-2 porcine circovirus. Can J Vet Res. 64:44–52. 2000.
13). Hall TA. Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98. 1999.
14). Harding JCS, Clark EG. Recognizing and diagnosing postweaning multisystemic wasting syndrome (PMWS). Swine Health Prod. 5:201–203. 1997.
15). Higgins DG, Sharp PM. CLUSTRAL; a package for performing multiple sequence alignment on a microcomputer. Gene. 73:237–244. 1988.
16). Knell S, Willems H, Hertrampf B, Reiner G. Comparative genetic characterization of porcine circovirus type 2 samples from German wild boar populations. Vet Microbiol. 109:169–177. 2005.

17). Krakowka S, Ellis JA, MCneilly F, Gilpin D, Meehan B, Rings DM, McCullough K, Botner A, Nauwynck H, Charreyere C, Allan G. The pathogenesis of PCV-2-associated post weaning multisystemic wasting syndrome in swine. Proc of 4th Internatl Symp on Emerging and Re-emerging Pig Diseas. p143–147. 2003.
18). Kim JH, Lyoo YS. Genetic characterization of porcine circovirus-2 field isolates from PMWS pigs. J Vet Sci. 3:31–39. 2002.

19). Larochelle R, Morin M, Antaya M, Magar R. Identification and incidence of porcine circovirus in routine field cases in Quebec as determined by PCR. Vet Res. 145:140–142. 1999.
20). Larochelle R, Margar R, D'Allaire S. Genetic characterization and phylogenetic analysis of porcine circo-virus type 2 (PCV2) strains from cases presenting various clinical condition. Virus Res. 90:101–112. 2002.
21). Larochelle R, Margar R, D'Allaire S. Comparative serologic and virologic study of commercial swine herds with and without postweaning multisystemic wasting syndrome. Can J Vet Res. 67:114–120. 2003.
22). Lekcharoensuk P, Morozov I, Paul PS, Thangthumniyom N, Wajjawalku W, Meng XJ. Epitope mapping of the major capsid protein of type 2 porcine circovirus (PCV2) by using chimeric PCV1 and PCV2. J Virol. 78:8135–8145. 2004.

23). Lyoo KS, Kim JH, Park CK. Identification of porcine circovirus with genetic variation from lymph nodes collected in pigs with PMWS. Korean J Vet Res. 39:353–358. 1999.
24). Lyoo KS, Park YH, Park BK. Prevalence of porcine reproductive and respiratory syndrome virus, porcine circovirus type 2 and porcine parvovirus from aborted fetuses and pigs with respiratory problems in Korea. J Vet Sci. 2:201–207. 2001.

25). Mahe D, Blanchard P, Truong C, Arnauld C, Le Cann P, Cariolet R, Madec F, Albina E, Jestin A. Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J Gen Virol. 81:1815–1824. 2000.
26). Meehan BM, NcNeilly F, Todd D, Kennedy S, Jewhurst VA, Ellis JA, Hassard LE, Clark EG, Haines DM, Allen GM. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J Gen Virol. 79:2171–2179. 1998.

27). Nawagitgul P, Morozov I, Bolin SR, Harms PA, Sorden SD, Paul PS. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J Gen Virol. 81:2281–2287. 2000.

28). Renee L, Ronald M, Sylvie DA. Genetic characterization and phylogenetic analysis of porcine circovirus type 2 (PCV2) strains from cases presenting various clinical conditions. Virus Res. 90:101–112. 2002.
29). Rosell C, Segales J, Plana-Duran J, Balasch M, Rodriguwz-Arrioja GM, Kennedy S, Allan GM, McNeilly F, Latimer KS, Domingo M. Pathological, immunohistochemical, and in-situ hybridization studies of natural cases of postweaning multisystemic wasting syndrome (PMWS) in pigs. J Comp Pathol. 120:59–79. 1999.

30). Sandvik T, Grierson S, King DP, Spencer Y, Banks M, Drew T. Detection and genetic typing of type 2 porcine circoviruses in archived pig tissues from the UK. Arch Virol. 149:1171–1183. 2004.
31). Segales J, Domingo M. Postweaning multisystemic wasting syndrome (PMWS) in pigs. A review. Vet Quart. 24:109–124. 2002.
32). Segales J, Rosell C, Domingo M. Pathological findings associated with naturally acquired porcine circovirus type 2 associated disease. Vet Microbiol. 98:137–149. 2004.

33). Tischer I, Mields W, Wolff D, Vagt M, Greim W. Studies on epidemiology and pathogenicity of porcine circovirus. Arch Virol. 91:271–276. 1986.

34). Tischer I, Gelderblom H, Vetterman W, Koch M. A very small porcine virus with circular single-stranded DNA. Nature. 295:64–66. 1982.

35). Wellenberg GJ, Stockhofe-Zurwieden N, de Jong MF, Boersma WJ, Elbers AR. Excessive porcine circo-virus type 2 antibody titers may trigger the development of porcine dermatitis and nephrophaty syndrome: a case-control study. Vet Microbiol. 99:203–214. 2004.
Figure 1.
PCR amplication for detection of PCV-2 ORF 2 gene. Lane 1; negative control (canine parvovirus), Lane 3; a negative sample, Lane 2 and 4; KOR-33 and KOR-29 using the primers PCV2F and PCV2B for 827 bp. Lane 5; KOR-71 using the primers 2S and 2AS for 496 bp. Lane M: 100 bp DNA marker (Bioneer, Korea).
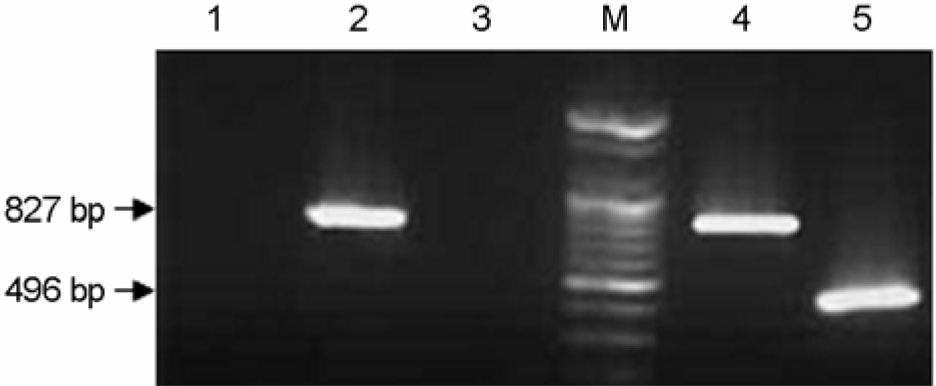
Figure 2.
Amino acid sequences alignments in three ORF2 immunorelevant epitopes of the PCV-2. Conserved sequences were shaded in three levels according to identity. A: The sequences (57 to 80 aa) were identical in the strains of KOR-94, KOR-8, KOR-80, GER NLPMWS 4, and TAI YL. B: The region of 121 to 136 aa reserved the identical situation of amino acids in KOR-71 and KOR-JHP strains, while the remaining 29 strains were different. C: The sequences (181 to 192 aa) were identical in the strains of KOR-71, US10489, US40895, KOR JHP, and KOR KSY-1.
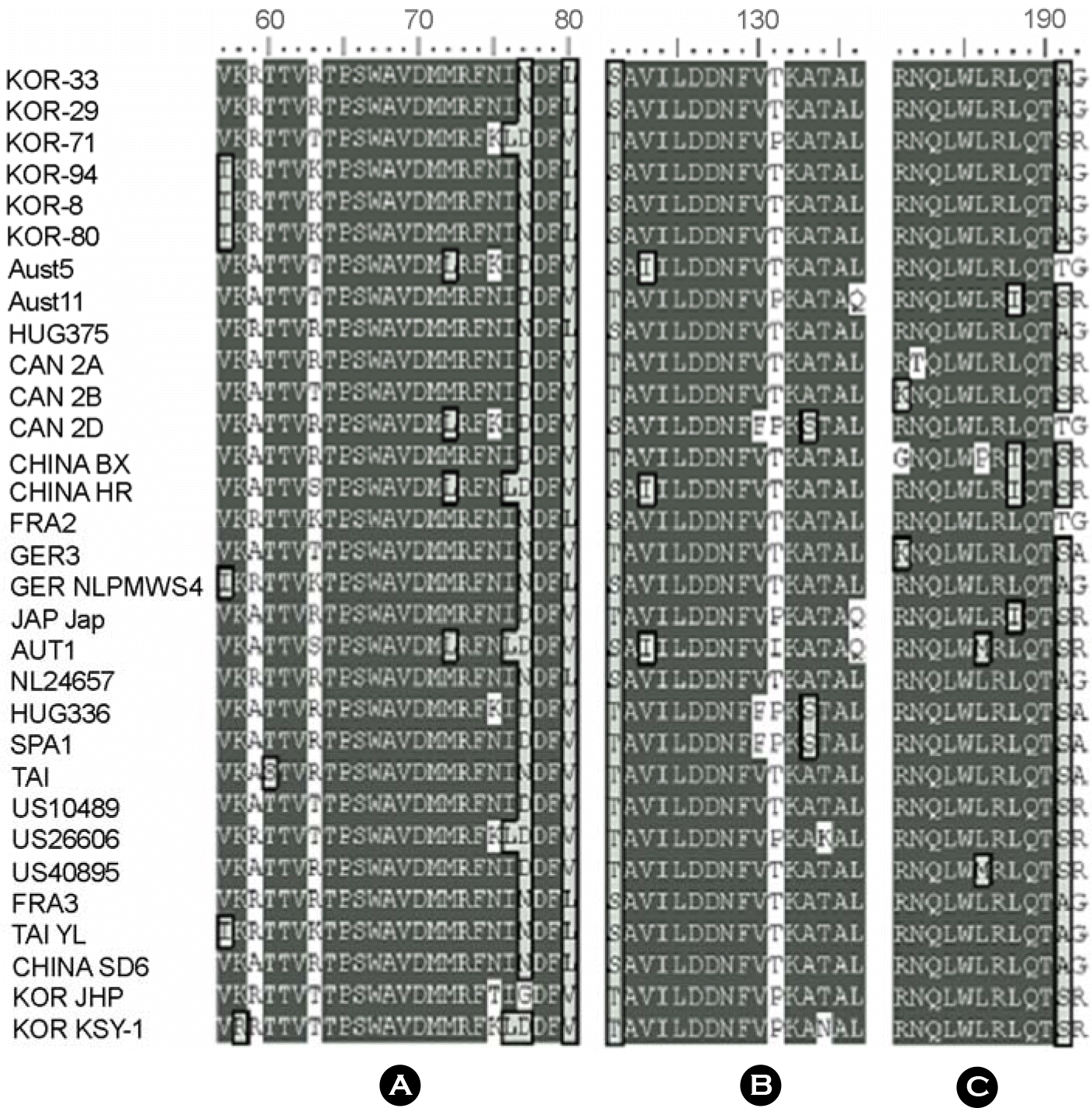
Figure 3.
Phylogenetic tree analysis of ORF2 gene (701 bp) of 31 strains of PCV-2. Six strains detected in this study, 2 Korean strains reported previously and 23 foreign strains were compared. The tree was constructed using computer analysis program Clustal X pakage. Bootstrap neighbor joining method was applied.
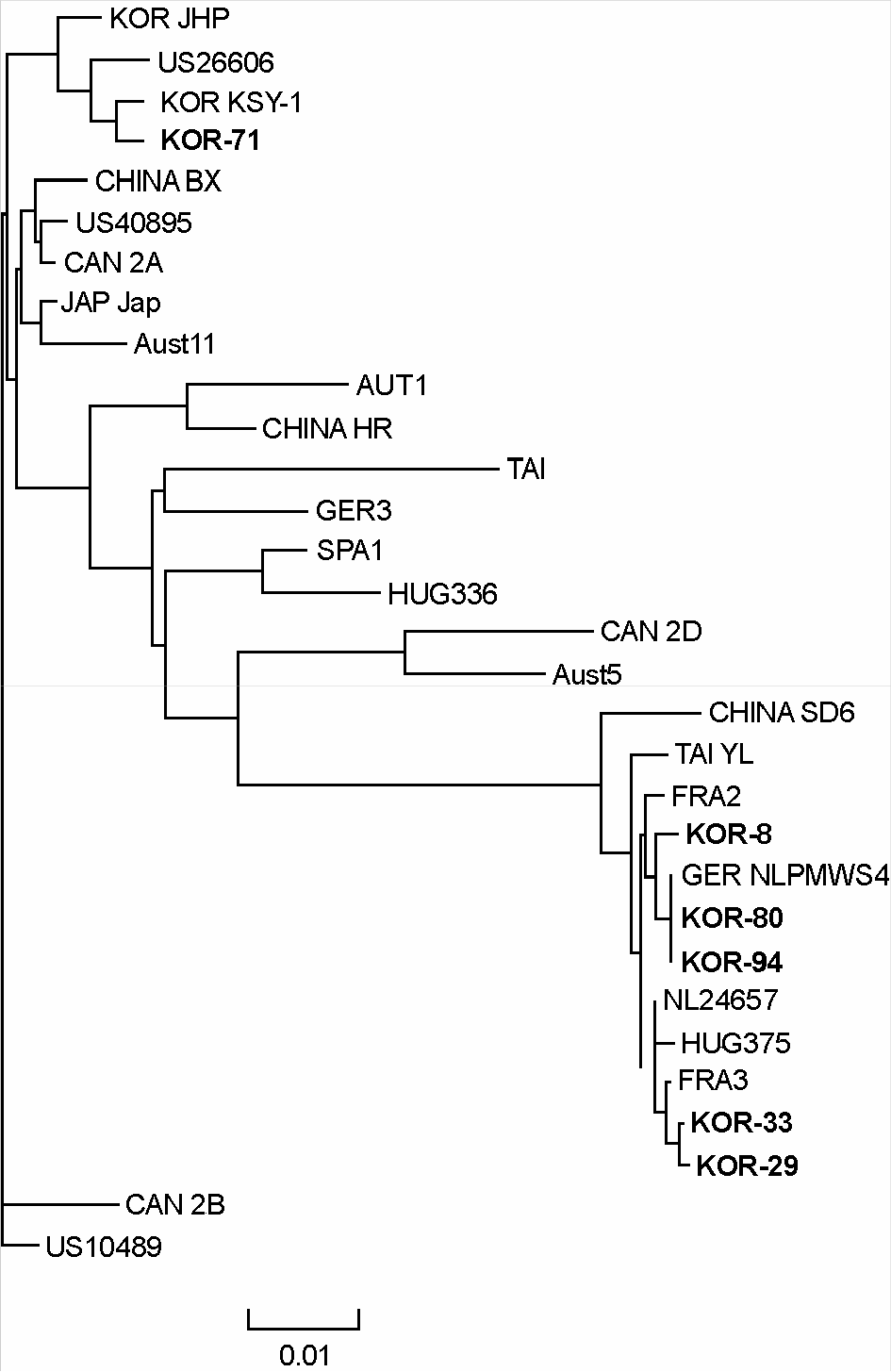
Table 1.
Sequences of oligonucleotide primers for PCR of PCV-2 ORF2 gene
| Primers | Primer sequence (5′→3′) | Size (nt) | Location (nt) |
|---|---|---|---|
| 2Sa | CAGTATATCCGAAGGTGCG | 19 | 1567∼1585 |
| 2AS | CATGGTTACACGGATATTG | 19 | 1081∼1103 |
| PCV2F | CCAGTTCGTCACCCTTTCC | 19 | 938∼956 |
| PCV2Bb | ACGAAAGAAGTGCGCTGTAAG | 21 | 1744∼1764 |
| PCVSa | ATGGGTCATAGGTTAGGGC | 19 | 1320∼1339 |
Table 2.
Summary of the swine herds for sample collection and PCR results
Table 3.
Comparison of the genomic and amino acid sequences of ORF2 (capsid protein) of PCV-2 strains identified in this study and other strains




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download