Abstract
Enteropathogenic Escherichia coli (EPEC) have been implicated in human diarrhea in several countries. Central to EPEC-mediated disease is its ability to cause intestinal lesions, known as attaching and effacing (A/E) lesion. We investigated 92 EPEC strains isolated from patients with diarrhea in Gwangju for their genotypic and phenotypic characteristics. Sixteen (17.4%) of all strains were found to be typical EPEC because they were bfpA gene positive by PCR. The most of typical EPEC isolates (87.5%) showed a localized adhesion (LA) pattern in Hep-2 cell adherence assay, whereas, only 11 atypical EPEC isolates (14.5%) were adhered to Hep-2 cells in a localized manner. Thirteen of the EPEC strains studied belonged to classical O-serogroups of EPEC and 7 isolates were classified as nonclassical EPEC serogroup and the other isolates could not be serotyped with our antisera. The subtypes of eae, tir, espA and espB genes which are major virulence genes concerned of A/E lesion on chromosome were analyzed by multiplex PCR for finding the original resource. The results showed that the composition of these genes subtypes was homogenous and heterogenous in 12 and 26 isolates, respectively. The others were non-determined type in terms of the gene subtype because of genetic diversity of intimin-coding eae genes. Our findings indicated that EPEC isolates from patients with diarrhea were diverse genetically and phenotypically, which require further study in regard to their virulence and epidemiological significance.
References
1). Agin TS, Wolf MK. Identification of a family of intimins common to Escherichia coli causing attaching-effacing lesions in rabbits, humans and swine. Infect Immun. 65:320–326. 1997.
2). China B, Goffaux F, Pirson V, Mainil J. Comparison of eae, tir, espA and espB genes of bovine and human attaching and effacing Escherichia coli by multiplex polymerase chain reaction. FEMS Microbiol Lett. 178:177–182. 1999.
3). Cravioto A, Gross RJ, Scotland SM, Rowe B. An adhesive factot found in strains of Escherchia coli belonging to the traditional infantile enteropathogenic serotypes. Curr Microbiol. 3:95–99. 1979.
4). Drolet R, Fairbrother JM, Harel J, Helie P. Attaching and effacing and enterotoxigenic Escherchia coli associated with enteric colibacillosis in the dog. Can J Vet Res. 58:87–92. 1994.
5). Eliane BN, Halha OS, Kinue L, Jacinta SP. Genotypic and phenotypic characterization of attaching and effacing Escherichia coli (AEEC) isolated from child with and without diarrhoea in Londria, Brazil. J Med Microbiol. 52:499–504. 2003.
6). Franke J, Franke S, Schmidt H, Schwarzkopf A, Wieler H, Baljer G, Beutin L, Karch H. Nucleotide sequence analysis of enteropathgentic Escherichia coli (EPEC) adherence factor probe and development of PCR for rapid detection of EPEC harboring virulence plasmids. J Clin Microbiol. 32:2460–2463. 1994.
7). Frankel G, Phillips AD, Rosenshine I, Dougan G, Kaper JB, Knutton S. Enteropathogenic and enterohemorrhagic Escherichia coli.; more subversive element. Mol Microbiol. 30:911–921. 1998.
8). Goffaux F, China B, Janssen L, Mainil J. Genotypic characterization of enteropathogenic Escherichis coli (EPEC) isolated in Belgium from dogs and cats. Res Microbiol. 151:865–871. 2000.
9). Gomez-Duartz OG, Kaper JB. A plasmid-encoded regulatory region activates chromosomal eae expression in entero-pathogenic Escherichia coli. Infect Immun. 63:1767–1776. 1995.
10). Gordillo ME, Reeve GR, Pappas J, Mathewson JJ, DuPunt HL, Murray BE. Molecular characterization of strains of enteroinvasive Escherichia coli O143, including isolates from a alrge outbreak in Houston, Texas. J Clin Microbiol Rev. 30:889–893. 1992.
11). Gunzberg ST, Tornieporth NG, Riley LW. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J Clin Micribiol. 33:1375–1377. 1995.
12). Kaper JB. Defining EPEC. Rev Microbiol Sao Paulo. 27:130–133. 1996.
13). Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 91:511–520. 1997.
14). Knutton S, Baldwin T, Williams PH, McNeish AS. Role of plasmid-encoded adherence factor in adhesion of enteropathogenic Escherichia coli to Hep-2 cells. Infect Immun. 55:78–85. 1987.
15). Kreuse G, Zimmermann S, Beutin L. Investigation of domestic animals and pets as a reservoir for intimin- (eae) gene positive Escherichia coli types. Vet Microbiol 20. 106(1–2):87–95. 2005.
16). Levine MM, Edelman R. Enteropathogenic Escherichia coli of classic serotypes associated with infant diarrhea: epidemiology and pathogenesis. Epidemiol Rev. 6:31–51. 1984.
17). Levine MM, Nataro JP, Karch H, Baldini MM, Kaper JB, Black RE, Clements ML, O'Brien AD. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J Infect Dis. 152:550–559. 1985.
18). Levine MM, Pardo V, Robins-Broene R, Lior H, Kaper JB, Moseley SL, Gicquelais K, Nataro JP, Vial P, Tall B. Use of DNA probes and HEp-2 cell adherence assay to detect diarrheagenic Escherichia coli. J Infect Dis. 158:224–228. 1988.
19). Mani R, Udgaonkar U, Pawar S. IStudy of enteropathogenic Escherichia col i(EPEC) diarrhoea in children. Indian J Pathol Microbiol. 46(1):118–20. 2003.
20). Marilda CV, Renata KTK, Angela MGD. Unidentified serogroup of enteropathogenic Escherichia coli (EPEC) associated with diarrhoea in infants in London, Parana, Brazil. J Med Microbiol. 49:823–826. 2000.
21). Moon HW, Whipp SC, Argenzio RA, Levine MM, Giannella RA. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect Immun. 41:1340–1351. 1983.
22). Nunes EB, Saridakis HO, Lrino K, Pelayo JS. Genotypic and phenotypic characterization of attaching and effacing Escherichia coli (AEEC) isolated from children with and without diarrhoea in Londrina, Brazil. J Med Microbiol. 52:499–504. 2003.
23). Scotland SM, Smith HR, Rowe B. Escherichia coli O128 strains from infants with diarrhea commonly show localized adhesion and positivity in the fluorescent-actin straining test but do not hybridize with an enteropathogenic E. coli adherence factor probe. Infect Immun. 59:1569–1571. 1991.
24). Vieira MAM, Andrade JRC, Trabulsi LR, Rosa ACP, Dias AMG, Rames SRT, Frankel G, Gomes TAT. Penotypic and genotypic characteristics of Escherichia coli strains of non-Enteropathogenic E. coli (EPEC) serogroups that carry eae and lack the EPEC adherence factor and shiga toxin DNA probe sequence. J Infect Dis. 183:762–772. 2001.
25). Viljanen MK, Peltola T, Junnila OL, Jarvinen H, Kuistila M, Huovinen P. Outbreak of diarrhoea due to Escherichia coli O111:B4 in schoolchildren and adults: association of Vi antigen-like reactivity. Lancet. 336:831–834. 1990.
26). Zhu C, Harel J, Jacques M, Desautels C, Donnenberg MS, Beaudry M, Fairbrother JM. Virulence properties and attra-ching and effacing activity of Escherichia coli O45 from swine post-weaning diarrhea. Infect Immun. 62:4153–4159. 1994.
Figure 1.
Adherence patterns in HEp-2 cells presented by EPEC isolates. A-1 and A-2, LA patterns (localized adherence). B-1 and B-2, AA patterns (aggregative adherence). C-1 and C-2, DA pattern (diffuse adherence). Original magnification, ×1000.
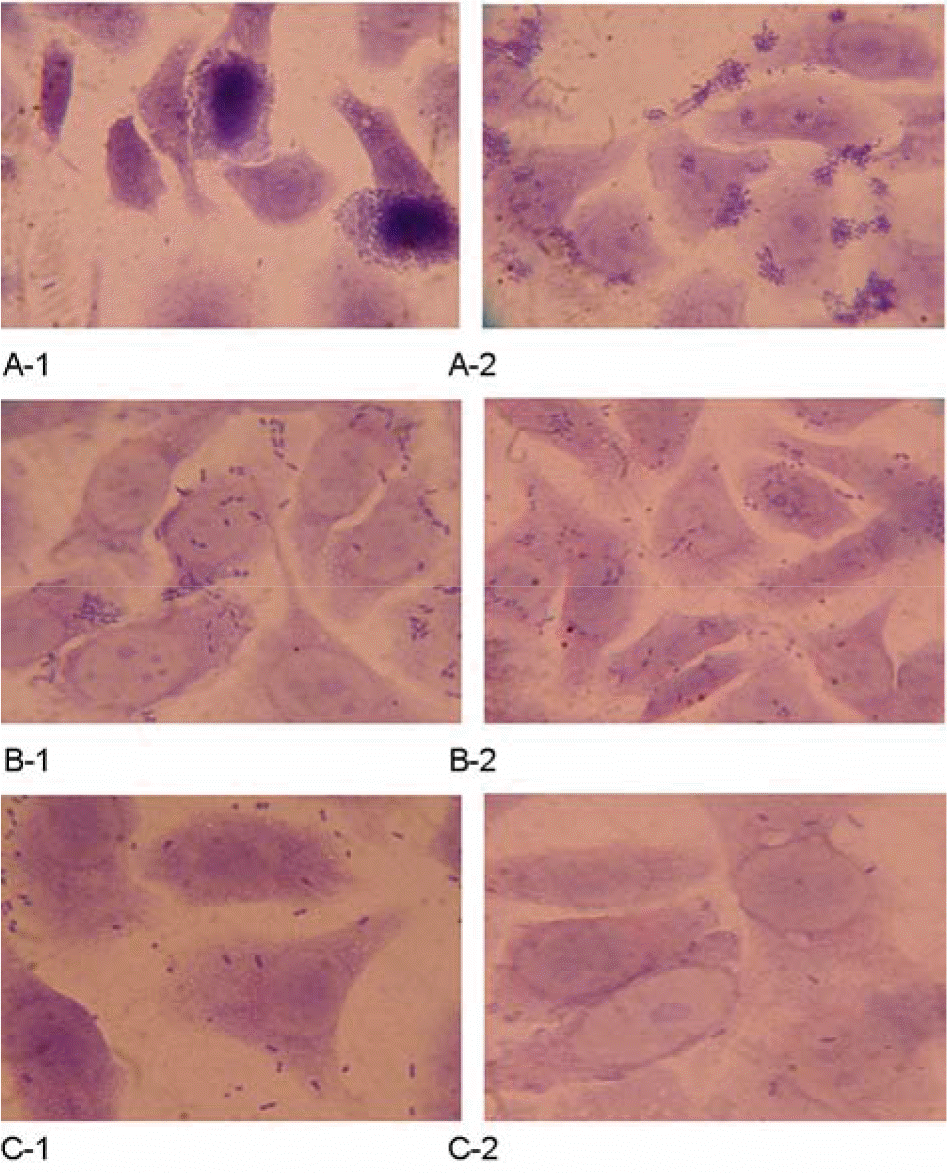
Figure 2.
PCR result to detection of bfpA and EAF gene. A, lane 1: 100 bp ladder; lane 2: bfpA+ isolate; lane 3: bfpA– isolate. B, lane 1: 100 bp ladder; lane 2: EAF+ isolate; lane 3: EAF– isolate.
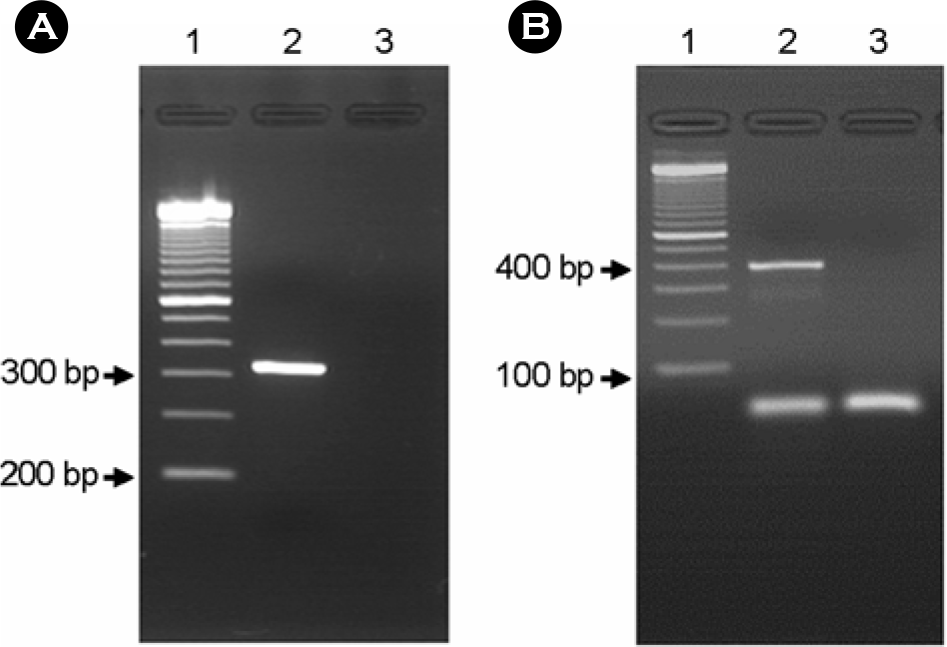
Figure 3.
Result of multiplex PCR to analysis subtypes of eae, tir, espA and espB genes. A, lane 1: 100 bp ladder; lane 2: eaeα; lane 3: eaeβ; lane 4: eaeγ. B, lane 1: 100 bp ladder; lane 2: tirα: lane 3: tirβ; lane 4: tirγ; lane 5: negative control. C, lane 1: 100 bp ladder lane 2: espAα; lane 3: espAβ, lane 4: negative control. D, lane 1: 100 bp ladder; lane 2: espBα; lane 3: espBβ; lane 4: espBγ; lane 5: negative control.
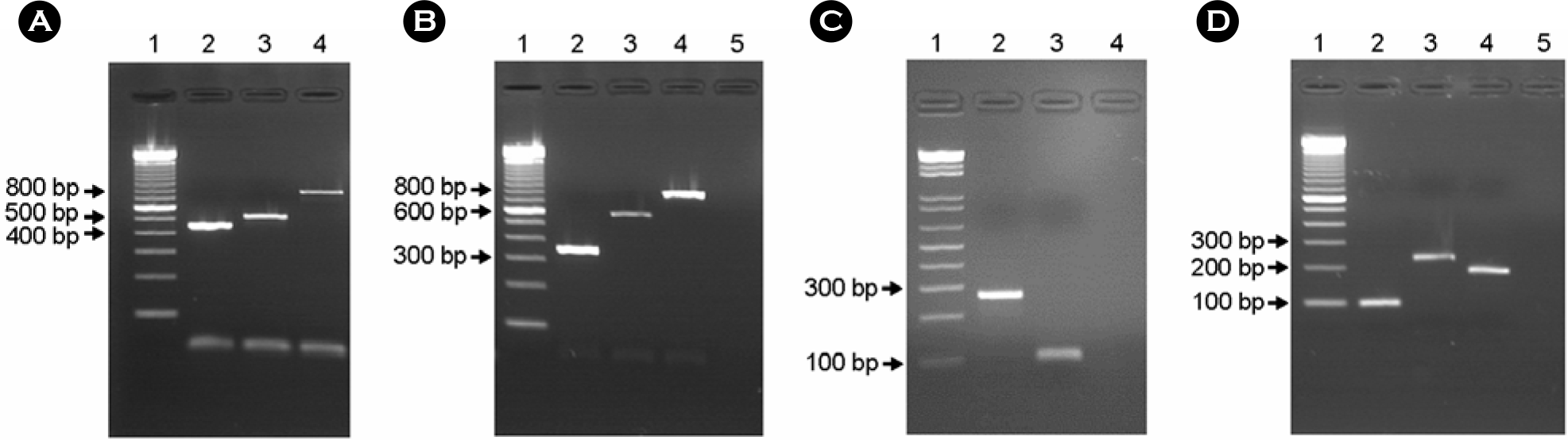
Table 1.
Primers used to detect for bfpA and EAF gene
Table 2.
Primers used to typing eae, tir, espA and espB gene
| Primer | Sequence of primer (5′ → 3′) | Target sequence | Reference strain | Product size (bp) |
|---|---|---|---|---|
| B73 | F: ACTGAGATTAAGGCTGATAA | conserved region of eae | ||
| B138 | R: GACCAGAAGAAGATCCA | eae α type | E2348/69 | 452 |
| B74 | R: AGGAAGAGGGTTTTGTGTT | eae γ type | EDL933 | 778 |
| B137 | R: TGTSGTCGCACTCTGATT | eae β type | 193 | 520 |
| B139 | F: CR∗CCK∗∗CCAY∗∗∗TACCTTCACA | conserved region of tir | ||
| B152 | R: CGCTAACCTCCAAACCATT | tir α type | E2348/69 | 342 |
| B141 | R: GTCGGCAGTTTCAGTTTCAC | tir γ type | EDL933 | 781 |
| B140 | R: GATTTTTCCCTCGCCACTA | tir β type | 95ZGI | 560 |
| B163 | F: TGAGGCATCTAARGM∗∗∗∗GTC | conserved region of espA | ||
| B165 | R: GCTGGCTATTATTGACCG | espA α type | E2348/69 | 269 |
| B164 | R: ATCACGAATACCAGTTACCA | espA γ type | EDL933 | 172 |
| B166 | R: GCCGTTTTTGAGAGCCA | espA β type | RDEC-1 | 101 |
| B148 | F: GCCGTTTTTGAGAGCCA | conserved region of espB | ||
| B151 | R: TCCCCAGGACAGCAGCAFT | espB α type | E2348/69 | 94 |
| B150 | R: GCACCAGCAGCCTTTGA | espB γ type | EDL933 | 188 |
| B149 | R: CTTTCCGTTGCCTTAGT | espB β type | RDEC-1 | 233 |
Table 3.
Genotypic and phenotypic characteristics of EPEC isolates
| Genetic profile (number of isolates) | HEp-2 cell adherence pattern | Type of | Serogroup (number of isolates) | |||
|---|---|---|---|---|---|---|
| eae | tir | espA | espB | |||
| eae, bfp, eaf (1) | LA (1) | NT∗ | α | α | α | OUT∗∗(1) |
| eae, bfp (15) | LA (13) | α | α | α | α | O157(1), O142(3) |
| NT | α | α | α | O114(2), O125(1), O157(1), OUT(4) | ||
| NT | α | NT | α | O157(1) | ||
| AA (2) | NT | α | α | α | OUT(2) | |
| eae (66) | LA (11) | α | α | α | α | OUT(1) |
| β | α | α | α | OUT(2) | ||
| NT | α | α | α | OUT(2) | ||
| NT | β | β | β | OUT(2) | ||
| NT | γ | α | α | O63(1), OUT(1) | ||
| NT | γ | β | γ | OUT(1) | ||
| NT | NT | α | α | OUT(1) | ||
| AA (43) | β | α | α | α | OUT(9) | |
| β | β | β | β | OUT(3) | ||
| γ | α | α | α | OUT(5) | ||
| γ | γ | β | γ | OUT(1) | ||
| NT | α | α | α | O114(4), OUT(8) | ||
| NT | β | β | β | O63(1), O157(1), OUT(4) | ||
| NT | β | β | NT | OUT(1) | ||
| NT | α | NT | α | O20(1), OUT(4) | ||
| NT | γ | β | γ | OUT(1) | ||
| DA (22) | β | β | β | β | O119(1), O128(1), OUT(2) | |
| γ | α | α | α | O128(1), OUT(8) | ||
| NT | α | α | α | OUT(7) | ||
| NT | β | β | β | OUT(2) | ||




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download