Abstract
Japanese encephalitis virus (JEV) is a member of mosquito-borne flaviviruses. To investigate whether there is a cis-acting genetic element in the coding region of the JEV C protein, which is required for viral replication, we generated four mutants by introducing a various size of deletions in each structural protein-coding region, designated as pJEV/Rep/ΔCC/LUC, pJEV/Rep/ΔC/LUC, pJEV/Rep/ΔprM/LUC, and pJEV/Rep/ΔE/LUC, of these, all replicons except for pJEV/Rep/ΔCC/LUC were competent in replication. Since pJEV/Rep/ΔCC/LUC is the same as pJEV/Rep/ΔC/LUC except for an additional 5′ deletion (nt 132∼201) in the coding region of the C protein, this region appeared to be essential for RNA replication. This is consistent with the proposed cyclization sequence motif in the 5′ region of the C gene, which has been recently shown to be required for replication in other mosquito-borne flaviviruses such as DV, YFV, KUN, and WNV. Thus, our results suggest that a cis-acting genetic element in the coding region of the JEV C protein may play an important role in RNA replication. This study will facilitate the current understanding of JEV RNA replication.
Go to : 
References
1). Ackermann M, Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926–39937. 2001.

2). Alvarez DE, Lella Ezcurra AL, Fucito S, Gamarnik AV. Role of RNA structures present at the 3UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 339:200–212. 2005.

3). Blumenthal T, Carmichael GG. RNA replication: function and structure of Qbeta-replicase. Annu. Rev. Biochem. 48:525–548. 1979.

4). Burke DS, Monath TP. Flaviviruses. p. 1043–1125. In. Knipe D.M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B, Straus S. E., editors(ed.).Fields virology. 4th ed.vol. 1. Lippincortt Williams & Wilkins;Philadelphia, Pa: 2001.
5). Cahour A, Pletnev A, Vazielle-Falcoz M, Rosen L, Lai CJ. Growth-restricted dengue virus mutants containing deletions in the 5′ noncoding region of the RNA genome. Virology. 207:68–76. 1995.

6). Chambers TJ, Halm CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649–688. 1990.

7). Contreras R, Cherouter H, Degrave W, Fiers W. Simple, efficient in vitro synthesis of capped RNA useful for direct expression of cloned eukaryotic genes. Nucleic Acids Res. 10:6353–6362. 1989.
8). Guirakhoo F, Monath TP. Immunoginicity, genetic stability, and protectiver efficacy of a recombinant, chimeric yellow fever-japanese encephalitis virus (chimerivax-JE) as a live, attenuated vaccine candidate against Japanese encephalitis. Virology. 257:363–372. 1999.
9). Hahn CS, Hahn YS, Rice CM, Lee E, Dalgarno L, Strauss EG, Strauss JH. Conserved elements in the 3untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198:33–41. 1987.
10). Herold J, Andino R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell. 7:581–591. 2001.

11). Hewlett MJ, Pettersson RF, Baltimore D. Circular forms of Uukuniemi virion RNA: an electron microscopic study. J. Virol. 21:1085–1093. 1977.

12). Hofacker IL, Fekete M, Flamm C, Huynen MA, Rauscher S, Stolorz PE, Stadler PF. Automatic detection of conserved RNA structure elements in complete RNA virus genomes. Nucleic Acids Res. 26:3825–3836. 1998.

13). Hofacker IL, Fontana W, Stadler PF, Bonhoeffer S, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structure. Monatsh. Chem. 125:167–188. 1994.
14). Hofacker IL, Stadler PF. Automatic detection of conserved base pairing patterns in RNA virus genomes. Comput. Chem. 23:401–414. 1999.

15). Holden KL, Harris E. Enhancement of dengue virus translation:role of the 3untranslated region and the terminal 3stem-loop domain. Virology. 329:119–133. 2004.
16). Hsu MT, Parvin JD, Gupta S, Krystal M, Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc. Natl. Acad. Sci. USA. 84:8140–8144. 1987.

17). Khromykh AA, Meka H, Guyatt KJ, Westaway EG. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719–6728. 2001.

18). Komar N, Savage HM, Stone W, McNamara T, Gubler DJ. Origin of the west nile virus responsible for an outbreak of encephalitis in the northeastern United states. Science. 286:2333–2337. 1999.

19). Lo MK, Tilgner M, Bernard KA, Shi PY. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 77:10004–10014. 2003.
20). Mackenzie JM, Khromykh AA, Jones MK, Westaway EG. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology. 245:203–215. 1998.

21). Mandl CW, Ecker M, Holzmann H, Kunz C, Heinz FX. Infectious cDNA clones of tick-borne encephalitis virus European subtype prototypic strain Neudoerfl and high virulence strain Hypr. J. Gen. Virol. 78:1049–1057. 1997.

22). Markoff L. 5- and 3-noncoding regions in flavivirus RNA. Adv. Virus Res. 59:177–228. 2003.
23). Rice CM. Flaviviridae: The viruses and their replication, p.931–960. In B. N. Fields, D. M. Knipe and P. M. Howley (ed), Fields Virology, Third ed, vol. 1. Lippincott-Raven Publishers, Philadelphia. 1996.
24). Solomon T. Origin and evolution of Japanese encephalitis virus in southest asia. J. Virol. 77.5:3091–3098. 2003.
25). Thiel HJ, Plagemann PGW, Moennig V. Pestiviruses. p. 1059–1073. In. Field B.N., Knipe D.M., Howley P.M., editors(ed),. Fields Virology. Raven Press;New York, N.Y.: 1996.
26). Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 2:135–140. 1998.

27). Yun SI, Kim SY, Lee YM. Development and apolication of a reverse genetics system for Japanese encephalitis virus. J. Virol. 77(11):6450–6465. 2003.
Go to : 
 | Figure 1.Construction of JEV viral replicons. The structures of the JEV viral replicons are shown. Solid boxes (▪) indicate in-frame deletions that had been introduced into the genome of the infectious pJEV/FL/LUC construct. Four constructs, namely, pJEV/Rep/ΔCC/LUC, pJEV/Rep/ΔC/LUC, pJEV/Rep/ΔprM/LUC, and pJEV/Rep/ΔE/LUC, contain a single in-frame deletion in each structural gene of JEV. pJEV/Rep/ΔCC/LUC has a deletion that extends to the proposed cyclization sequence motif in the 5′ region of the C gene, unlike pJEV/Rep/ΔC/LUC. EMCV IRES, Internal ribosome entry site element of encephalomyocarditis virus; LUC, luciferase gene. |
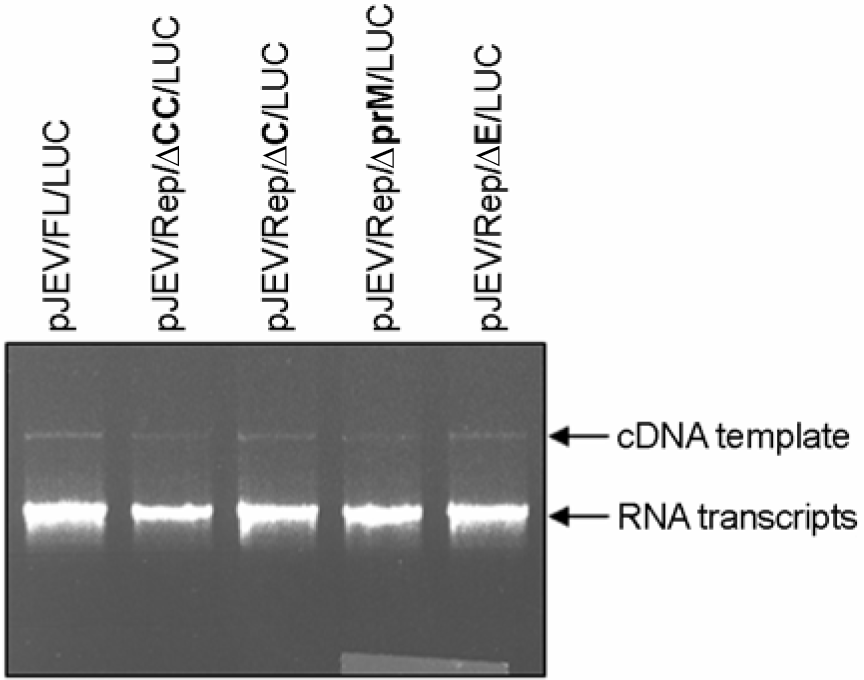 | Figure 2.Generation of the synthetic RNA transcripts in vitro transcribed from pJEV/Rep/ΔCC/LUC, pJEV/Rep/ΔC/LUC, pJEV/Rep/ΔprM/LUC, and pJEV/Rep/ΔE/LUC cDNA templates. Each cDNA as indicated was linearized by XbaI digestion. Linearized cDNA templates were used for in vitro transcription reaction using the SP6 RNA polymerase. 1∼1.5 μl of the reaction mixtures was analyzed by 0.5% agarose gel electrophoresis. cDNA templates and RNA transcripts as indicated were visualized by ethidium bromide staining. |
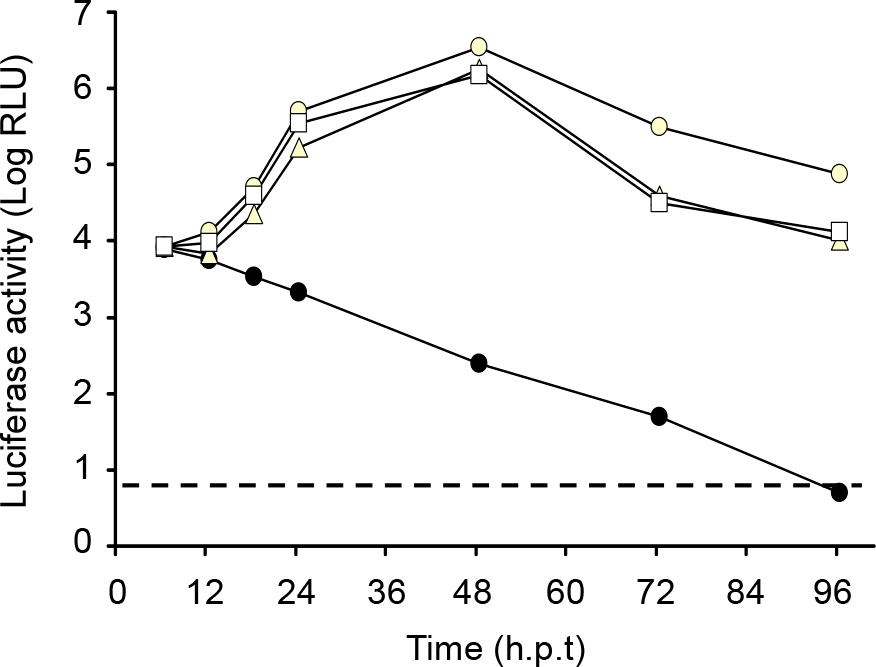 | Figure 3.Expression of LUC. Naive BHK-21 cells (8×106) were transfected with 2 μg of the parent or JEV viral replicon RNAs that had been transcribed from each cDNA template and then seeded on 6-well plates at a density of 4×105 cells per well. At the indicated time points, the cell lysates were subjected to luciferase assays. The experiments were performed in triplicate and the mean values are shown. Solid circle, pJEV/Rep/ΔCC/LUC; open circle, pJEV/Rep/ΔC/LUC; open triangle, pJEV/Rep/ΔprM/LUC; open square, pJEV/Rep/ΔE/LUC. A dotted line indicates the level of background luminescence of naïve cells. hpt, hours post-transfection. |
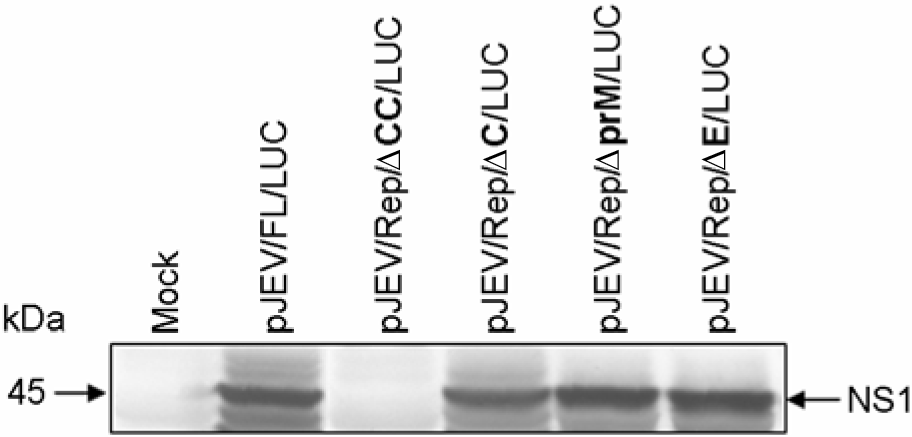 | Figure 4.Expression of JEV NS1 protein. The transfected cells (4×105) as indicated were lysed with 1X sample loading buffer 48 hr post-transfection and the protein extracts were resolved on 10% SDS-polyacrylamide gels. The proteins were transferred onto the nitrocellulose membrane and immunoblotted with a JEV hyperimmune antiserum. |
Table 1.
Oligonucleotides used in this study
Table 2.
플라비바이러스의 cyclization sequence




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download