Abstract
Background
Foot-and-mouth disease virus (FMDV) is a small single-stranded RNA virus which belongs to the family Picornaviridae, genus Apthovirus. It is a principal cause of FMD which is highly contagious in livestock. In a wild type virus infection, infected animals usually elicit antibodies against structural and non-structural protein of FMDV. A structural protein, VP1, is involved in neutralization of virus particle, and has both B and T cell epitopes. A RNA-dependent RNA polymerase, 3D, is highly conserved among other serotypes and strongly immunogenic, therefore, we selected VP1 and 3D as vaccine targets.
Methods
VP1 and 3D genes were codon-optimized to enhance protein expression level and cloned into mammalian expression vector. To produce recombinant protein, VP1 and 3D genes were also cloned into pET vector. The VP1 and 3D DNA or proteins were co-immunized into 5 weeks old BALB/C mice.
Foot-and-mouth disease (FMD) is a lethal vesicular disease in cloven-hoofed animals, and it infects lung epithelial cells in respiratory tract. It is transmitted by air and develops blisters rupture, and pyrexia on mouth and feet. It is caused by Foot-and-mouth disease Virus (FMDV) which is a member of the family Picornaviridae, genus Apthovirus, having 7-8 kb positive single-stranded RNA genome (1). Seven immunologically distinct serotypes have been identified on the basis of a VP1 coding region sequence: A, O, C, SAT1, SAT2, SAT3, and Asia1. Serotype O is prevalent in Africa, Asia, South America, and occasionally Europe. It accounts for over 60% of positive FMD isolated by The Food and Agriculture Organization World Reference Laboratory for Foot-and-Mouth Disease (WRLFMD) from 2000 to 2004 (2).
Chemically inactivated FMDV is used as a commercial vaccine. In the early time, formalin and aziridine compounds were used widely for inactivation, however, they had a safety problem. Later, binary ethyleneimine (BEI) was found to be as a more effective reagent, however, it was known to reduce efficacy of vaccination (3). Nowadays, many researchers have tried to use alternative vaccines which are more safe and effective such as a DNA vaccine for replacing these conventional vaccines.
In case of FMDV, structural protein genes are used as the most immunodominant target for DNA vaccine. A highly conserved Arg-Gly-Asp (RGD) triplet motif, which is located on the highly mobile exposed G-H loop of capsid protein VP1, has been reported as a neutralizing epitope site on empty FMDV capsids which is generated from in vivo infection. Sites 135~167 and 141~160 region (G-H loop) are known as the T and B cell epitopes. Moreover, 200~213 region of carboxyl terminus and 43~44 region of N-terminal also contain B cell epitopes (4,5). An expression of MHC class I complex were suppressed in 2~3 hr after virus infection, because virus eliminates translation initiation factor in host cells such as macrophage or dendritic cell in vitro (6). Although VP1 sequence is highly variable among serotypes and VP1 alone can't induce neutralizing antibody (7), DNA vaccine of VP1 can protect mice from viral infection without induction of neutralizing antibody (8). Nevertheless, VP1 is less likely to induce FMDV-specific T cell response than P1, because it is susceptible to proteolytic cleavage. Viral structural proteins have a tendency to induce humoral response, whereas nonstructural proteins seem to be more effective in inducing cellular immunity. A 3D, RNA-dependent RNA polymerase has a consistent sequence among various serotypes and is known as a stimulator of cellular and humoral immune response (9). Furthermore, it induces FMDV-specific T cell proliferation and delayed-type hypersensitivity (DTH) responses in pigs when DNA vaccine with P1 is introduced (10).
Intradermal injection using tattoo device was reported as an effective delivery system for induction of cellular immunity against viral infection (11,12). Moreover, antigen delivery through skin is also expected to be effective inducer of the innate immune response, because the antigen presenting molecules on the surface of swine skin dendritic cell (DC), such as swine major histocompatibility complex class II (SLA II) or co-stimulatory molecule CD80/ CD86, are not influenced by FMDV infection (13).
In this study, codon-optimized VP1 and 3D DNA vaccines as well as bacterial recombinant proteins VP1 and 3D were evaluated for their efficacy in mice, as determined by Ab ELISA and IFN-γ ELISpot assay.
RD and 293T cells were grown in Dulbecco's modified Eagle medium (DMEM, Gibco-BRL, Eggenstein, Germany) containing 10% heat-inactivated fetal bovine serum (FBS, Sigma, St. Louis, US).
VP1 and 3D of FMDV serotype O/otaiwan97 sequence were codon optimized for increasing protein expression level. PCR primers for each different VP1 and 3D were designed. These genes were amplified by conventional PCR amplification procedure and reaction condition. PCR included 30 cycles of denaturation at 94℃ for 1 min, annealing at 55℃ for 1 min, extension at 72℃ for 1 min, and final extension at 72℃ for 10 min. Amplified PCR products were cloned into pGEM-T easy vector (Promega, Madison, USA), and sequences of the inserts were confirmed by sequencing analysis. Confirmed constructs were subcloned into pcDNA3.1 His/V5 (Invitrogen, Carlsbad, USA) and pET32a(+) bacterial expression vector (Novagen, Madison, USA). The primer sequences used for pcDNA3.1-VP1 cloning were forward 5'-GCCCCCAAGCTTGCCGCCACCATGACCAVVTCTGCTGGTGAG-3' and reverse 5'-ATCGGGCTCGAGTTTTGCAGGTGCCAC-3'. The primers used for pcDNA3.1-3D amplification were forward 5'-GCCCCCAAGCTTGCCGCCACCATGGGTTTGATCGTCGATACC-3' and reverse 5'-ATCGGGCTCGAGCGCGTCACCGCACACGG-3'. The VP1 used for cloning into pET32a-VP1 was amplified by PCR. The sequence of the sense and antisense primer were 5'-GCCCCCGGATCCACCACCTCTGCTGGTGAG-3' and 5'-ATCGGGAAGCTTTTTTGCAGGTGCCAC-3', respectively. The primers used for pET32a-3D amplification were 5'-GCCCCCGGATCCGGTTTGATCGTCGATACC-3' for forward and 5'-ATCGGGAAGCTTCGCGTCACCGCACACGG-3' for reverse. For DNA immunization, each DNA plasmid was amplified in E. coli strain DH5α (Gibco-BRL, Bethesda, USA) and purified using an Endofree Plasmid Maxi kit (QIAGEN Inc, Valencia, USA).
Plasmids, pET32a-VP1 and pET32a-3D, were transformed into E. coli BL21-DE3 competent cells (Gibco-BRL, Gaithersburg, USA). The bacteria were cultured in 500 ml LB at 37℃ until OD600 reached 0.6. Expression was induced by the addition of isopropyl-β-thiogalactopyranoside (IPTG) at a final concentration of 0.5 mM and incubated at 37℃ for 3 hrs for 3D or 6 hrs for VP1. The cells were harvested and lysed by sonication on ice, followed by centrifugation. The supernatant was used for purification of 3D, while the pellet of VP1 was solubilized with EDTA-free binding buffer (20 mM Tris-HCl, pH 7.9, 0.5 M NaCl, 8 M urea) and the cell debris was discarded. The protein of interest was isolated and purified using ProBond™ Columns (Invitrogen Corporation, Carlsbad, USA) by ProBond purification system with Ni-NTA resin (QIAGEN, Chatsworth, USA). Then, the proteins were eluted with a gradient of elution buffer (20 mM Tris-HCl, pH 7.9, 0.5 M NaCl, 1M imidazole). Eluted protein was stored at -80℃ until assay.
The assay was carried out according to the method previously described (14). RD cells at a density of 1.2×105 cells were subcultured in DMEM in 2-well chamber slide for 16 hrs before transfection. DNA was mixed with Fugene6 (Roche Molecular Biochemicals, Indianapolis, USA) and added dropwise to the cell. After 48 hrs of incubation, transfected cells were washed with serum-free medium and 1X phosphate buffered saline (PBS) once. Washed cells were fixed with 2% paraformaldehyde (Sigma Chemical Co., St. Louis, USA) solution for 30 min and washed three times with 1X PBS. After blocking with 10% goat serum in 0.1% Triton X-100 for 30 min, the cells were incubated with mouse anti-V5 monoclonal antibody (Invitrogen, Groningen, The Netherlands) at a 1:1,000 dilution in 0.1% Triton X-100 with 3% goat serum for 90 min at room temperature (RT). After washing three times with 1X PBS, cells were incubated with fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG antibody (Molecular Probe, Carlsbad, USA) at a 1:2,500 dilution in 0.1% Triton X-100 with 3% goat serum for 90 min at RT. The cells were washed three times with 1X PBS. Washed cells were incubated with 4'-6-Diamidino-2-phenylindole (DAPI, Roche, Indianapolis, US) for 15 min at RT, and they were then washed extensively in 1X PBS and a cover slip was mounted over the cells using mounting medium (Shandon, Pittsburg, US). The prepared slides were observed under UV microscope (Nikon, ECLIPSE TE2000-U, Tokyo, Japan).
Cells were harvested 48 hrs after transfection with CaPO4 precipitation using a ProFection Kit (Promega, Madison, WI) and lysed with lysis buffer (50 mM Tris-Cl, pH=7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, protease inhibitor cocktail). The cell lysate in 3 X SDS loading buffer was boiled for 5 min and electrophoresed through 10% SDS-PAGE gel. Resolved proteins were transferred to a nitrocellulose membrane and the membrane, was blocked with 5% milk protein in TBST (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.05% Tween 20) for 4 hrs at RT. The membrane was incubated with an anti-V5 monoclonal antibody (Invitrogen, Groningen, The Netherlands) in the blocking solution for 2 hrs at RT and washed in TBST. Washed membrane was incubated with HRP-conjugated anti-mouse IgG antibody (Molecular Probe, Carlsbad, USA) in the blocking solution for 1 hr at RT, washed, and developed using ECL kit (AbFrontier, Seoul, Korea).
Peptides for VP1 and 3D were synthesized on the basis of B cell epitope prediction and immunized into rabbits to raise VP1- and 3D-specific antisera. Rabbits were injected with 200 µg each of different peptides in Freund's complete adjuvant (FCA, Sigma, USA), followed by two boosting immunizations at 4-week intervals according to conventional immunization scheme.
Six to eights weeks old female BALB/c mice were divided into several groups (4 mice/group) for the DNA or protein immunization
Mice were immunized with total 10µg of purified protein via intramuscular (i.m.) route at 2 weeks interval. Group 1 was immunized with VP1 and 3D protein. Group 2 was immunized with PBS as a negative control. Mice were immunized 8 times from days 0. The splenocytes were harvested 3 weeks after the last boosting for analysis.
Mice were immunized 3 times in one cycle at 3 days interval with total 20µg of naked plasmid via i.d. route administration using tattoo device. Group 1 was immunized with pcDNA-VP1/-3D, which Group 2 was immunized with pcDNA3.1 empty vector as a negative control. Mice were immunized 6 cycles with 3 times per each cycle from days 0. The splenocytes were harvested 3 weeks after the last boosting for analysis.
An induction of antibodies in immunized mice was determined using an enzyme-linked immunosorbent assay (ELISA). A 96-well EIA/RIA plate (Corning Incorporated Costar, Lowell, MA, US) was coated with 5 µg/ml of purified protein in PBST (1X PBS, pH 7.4, 0.05% Tween-20) for 16 hrs at 4℃. The plates were blocked with 3% bovine serum albumin (BSA) in PBST for 1 hr at 37℃. After washing, 50 µl of sera (diluted 1:50) were added and incubated for 1 hr at 37℃. Bound antibodies were detected with HRP-conjugated anti-mouse IgG antibody and substrate TMB (3,3',5,5'-tetramethylbenzidine) buffer solution (Sigma, St. Louis, USA). Color reaction was stopped by adding 2M sulfuric acid, and an absorbance was measured at 450 nm using automated plate reader Bench mark plus system (Bio-Rad, Hercules, USA).
All reagents for ELISpot assay was purchased from BD Biosciences (Franklin Lakes, USA), unless otherwise specified. BD™ ELISOPT Plates were coated with 100 µl of Purified Anti-mouse IFN-γ antibody at a concentration of 5 µg/ml in sterile PBS overnight at 4℃. Coated plates were then washed once with 200 µl of RPMI 1640 (Sigma, St. Louis, USA) containing 10% FBS (Sigma, St. Louis, USA) and 1% Penicillin-Streptomycin (Gibco-BRL, Gaithersburg, USA), and then blocked with 200 µl of RPMI 1640 complete medium for 2 h at RT. The splenocytes were harvested from mice at one week after the last boosting, by grinding the spleen in RPMI 1640 containing 5% FBS and 1% pen/strep, and RBC was removed using Gey's medium. Cell suspension was centrifuged, and the cell pellet was resuspended in 10% RPMI 1640. Splenocytes were prepared at density of 4×105 cells/well in 10% RPMI medium, and they were stimulated with various peptides (10 µg/ml) for 48 hrs, and sequentially washed with deionized (DI) water, and PBS containing 0.05% Tween-20 for 3-5 min per each. One hundred µl of 2 µg/ml biotinylated anti-mouse IFN-γ in PBS containing 10% FBS was added to each well and incubated for 2 hrs at RT. Unbound antibody solution was discarded, and the precipitate was washed three times with 200 µl of washing buffer (PBS containing 0.05% Tween-20). One hundred µl of diluted streptavidin-HRP in PBS containing 10% FBS was added to per each well, and they were incubated for 1 hr at RT. And they were then washed 4 times with washing buffer for 1~2 min each, and were further washed twice with 200 µl of 1X PBS. Finally, 100 µl of Final Substrate Solution, BDTM AEC Substrate Reagent Set were added to each well. The plate was monitored for spot development from 60 min after incubation, so as not to be over-developed, and the reaction was stopped by washing wells with DI water. The plate was air-dried overnight at RT until being completely dried. Removal of plastic tray under plate will facilitate drying. The plate was stored in a sealed plastic bag in the dark until being analyzed. The number of spots per well was determined using a KS ELISPOT Automated Reader System with KS ELISPOT 4.2 Software (Carl Zeiss, Inc. Thornwood, USA).
Recombinant FMDV type O VP1 and 3D genes were generated by overlapping PCR as illustrated in Fig. 1A. After confirming sequence of DNA, the DNAs were ligated into pcDNA3.1 V5/His vector. The expression of the VP1 and 3D proteins in mammalian cells was determined by an IFA and Western blot analysis (Fig. 1). According to the Ag-specific signal, both VP1 and 3D proteins were found to the expressed in the cytoplasm of RD cells (Fig. 1B). However, no signals of pcDNA-transfected cells were detected. The 293T cell lysates of VP1 and 3D were harvested after 48 hrs of transfection, and were analyzed by Western blot analysis, using mouse anti-V5 antibody. As shown in Fig. 1C, the expected molecular weights of the pcDNA-VP1 and pcDNA-3D were 26.9 kDa and 56.2 kDa, respectively.
When both VP1 and 3D proteins were expressed in BL21-DE3 cells, only 3D protein was expressed as a soluble form. However, VP1 was expressed as an inclusion body and purified as a native form using hybrid purification method and used for a production of B-cell epitope-specific Abs. The VP1 and 3D protein bands on Coomassie stained gel were detected with a molecular weights, corresponding to 42 kDa (lane 1) and 72.8 kDa (lane 2), respectively (Fig. 2A). The VP1 and 3D B-cell epitope peptides were predicted, synthesized and immunized into rabbits. The purified proteins were confirmed by Western blot analysis using the VP1 or 3D B cell epitope peptide-specific polyclonal Abs which had been raised from rabbits. Fig. 2B shows that the recombinant VP1 and 3D proteins can be detected by the polyclonal Abs. The anti-B cell epitope polyclonal antibodies were used in ELISA for confirmation of activity.
Mice (BALB/c) were immunized with 10 µg of both pcDNAVP1 and pcDNA-3D plasmids, or 10 µg of both recombinant protein of VP1 and 3D. The mice sera were then harvested at 2 and 5 weeks post-immunization, and serum IgG levels were determined accoding to in vivo schedule (Fig. 3A). The levels of the humoral immune responses not only in the group co-immunized with VP1 and 3D DNA plasmid (Fig. 3B), but also in the group co-immunized with VP1 and 3D protein (Fig. 4B) showed higher titer of Ag-specific serum IgG than the mice immunized with pcDNA empty vector or PBS. The humoral immune responses were detected in the sera of all DNA- or protein-immunized groups as early as 2 weeks after the first immunization, and 3D-specific serum IgG level was increased dose-dependently in both 3D-DNA and protein immunized groups (Fig. 3B, 4B). IgG isotyping analysis with the sera at 9 weeks post-immunization showed that IgG2a/IgG1 ratio was slightly larger than 1. Therefore, VP1/3D DNA or protein vaccine seemed likely to induce Th1-dependent humoral immune response (Fig. 3C, 4C).
Cell-mediated immune response was determined by counting IFN-γ secreting cell number by ELISpot analysis. Fig. 5 shows that in DNA-immunized group, only 3D-specific IFN-γ secreting cells were stimulated, but not in VP1 or pcDNA. When VP1/3D proteins were co-immunized into mice, the IFN-γ secretion was antigen-specifically induced by both VP1 and 3D (Fig. 6).
In this study, recombinant DNA and protein vaccination approach was successfully achieved by co-administration of the FMDV type O VP1/3D, which contains both the B cell and T cell epitopes. The nucleotide sequences were codon-optimized, and its expression was confirmed in both mammalian cells and bacterial system. The purified proteins were found to be immunogenic and to have preserved B-cell epitope. In vivo study showed that VP1 and 3D-specific serum IgG were detected as early as 2 weeks after the first immunization and increased dose-dependently thereafter in both 3D-DNA and-protein immunized groups (Fig. 3, 4). Recombinant protein vaccine induced higher level of humoral immune response than naked DNA vaccine. Furthermore, the IgG2a/IgG1 ratio in the sera from the mice co-immunized with either pcDNA VP1/3D or recombinant VP1 and 3D protein was slightly higher than 1. In general, such pattern is typical in the Th1-dependent immune response. It was reported that DNA plasmid encoding P1 induces Th2-dependent immune response, whereas VP1 induces Th1-dependent humoral immune response (15). However, Park et al. showed that VP1 induces Th2-dependent immune response, when VP1 plasmid was coadministered with IL-1α via tail vein injection (16). Therefore, it seems to be important to choose molecular adjuvants to co-administer with VP1 for skewing immune responses. When the VP1 sequence which contains only B-cell epitope and non-structural protein (NSP) (17) or transgenic membrane-anchored VP1 (7) was immunized, the protection level was very low inspite of higher total Ab level. Soluble recombinant VP1 protein produced from E. coli also showed T-cell immune response and protection without generating neutralizing antibody (5). Furthermore, co-immunization of DNA plasmid encoding VP1 which contained all of the epitopes and NSP containing the T-cell epitope showed partial protection effect in guinea pig (18), or anti-viral protection effect irrespective of presence of neutralizing antibody titers (8).
ELISpot data detected IFN-γ secreting cells after stimulation with VP1 and 3D recall antigen, indicating that antigen-specific CD4+ T cell activation was induced in DNA or protein immunization (Fig. 5, 6). Only the mice immunized with VP1 DNA failed to stimulate the IFN-γ secreting cells when stimulated with recombinant VP1 protein (Fig. 5). It is, therefore, possible that the binding of VP1 RGD region to the cellular receptor interferes with the recognition of the T cell epitope (19). While VP1 induces humoral immune response, 3D induces cellular immune response more strongly.
In conclusion, the codon-optimized VP1, 3D DNA and recombinant proteins are strong immunogen to mice even with smaller amount. Especially, Ag-specific humoral and cellular immune responses were induced as early as 2 weeks post-immunization by using tattoo device. DNA vaccine is one of the better-defined subunit vaccine, and has better strategies to overcome many problems than protein vaccine, such as glycosylation, toxicity, cost or lack of the CTL activation (20). In order to compensate the weak and short-period immune responses generated by DNA vaccine, however, strong adjuvant molecules or powerful delivery tools are necessary. Our future research is aimed to construct both VP1 and 3D fusion plasmid and molecular adjuvant for induction of higher cellular immune response against VP1 and protective efficacy. Moreover, other delivery system such as electroporation would be worth to be considered. Finally, it is also necessary to evaluate protective immune response induced by DNA and protein vaccination, and elucidate the immune cells involved in anti-viral infection.
Figures and Tables
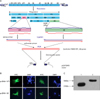 | Figure 1FMDV type-O VP1, 3D cloning strategy and in vitro protein expression. FMDV VP1 (0.6 Kb) and 3D (1.4 Kb) PCR products were subcloned into pcDNA3.1V5/His mammalian expression vector and pET bacterial expression vector. (A) FMDV type-O VP1 and 3D cloning strategy (B) Protein expression of VP1 and 3D was determined in plasmids-transected RD cells, and its expression was visualized by FITC-labeling and DAPI staining for nucleus. (C) 293T cells were transiently transfected with pcDNA-VP1 (lane 1) and pcDNA-3D (lane 2) plasmids, and the expression was confirmed by Western blot analysis. |
 | Figure 2Purification of recombinant VP1 and 3D proteins, and biological activity of B cell epitope peptide polyclonal Ab. (A) pET-VP1 (lane 1) and -3D (lane 2) expressions in bacterial system were confirmed by Coomassie staining of SDS-PAGE gel. (B) The purified VP1 (lane 1) and 3D (lane 2) proteins were confirmed by B cell epitope peptide -specific polyclonal Abs. |
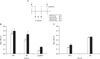 | Figure 3Humoral immune responses in DNA vaccine-immunized mice. (A) In vivo DNA immunization scheme (B) The antigen-specific serum IgG responses in Balb/c mice after co-immunization with pcDNA-VP1 and pcDNA-3D at two different time points (□ 2 wks p.i., ■ 5 wks p.i.). (C) IgG isotyping analysis with sera harvested from the group at 9 wks from the first immunization and tested for antibodies at 1:50 dilution. (□ IgG1, ■ IgG2a). The result was obtained from averages of groups against each antigen. The data represent average±S.D. |
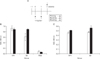 | Figure 4Humoral immune responses in protein vaccine-immunized mice. (A) In vivo protein immunization scheme (B) The antigen-specific serum IgG responses in Balb/c mice after immunization against VP1 or 3D protein at two different time points. (□ 2 wks p.i., ■ 5 wks p.i.) (C) IgG isotyping analysis with sera from the group sampled at 7 wks from the first immunization and tested for antibodies at 1:50 dilution (□ IgG1, ■ IgG2a). The result was obtained from averages of four mice in each group. The data represent average±S.D. |
 | Figure 5Cellular immune response in DNA vaccine-immunized mice. Numbers of IFN-γ secreting cell spots from a pool of 4 mice splenocytes (4×105 cells/well) were determined by recall Ag VP1 or 3D protein stimulation. The splenocytes were pooled from 4 mice harvested 3 weeks after the last vaccination with VP1 and 3D DNA. The ELISpot assay was performed after stimulating the cells with 10 µg/ml each of the recombinant proteins for 48 hrs. |
 | Figure 6Cellular immune responses in proteins vaccine-immunized mice. Numbers of IFN-γ secreting cell spots from a pool of 4 mice splenocytes (4×105 cells/well) were determined by recall Ag VP1 or 3D protein stimulation. The splenocytes were pooled from 4 mice harvested 3 weeks after the last vaccination with VP1 and 3D protein. The ELISpot assay was performed after stimulating the cells with 10 µg/ml each of the recombinant proteins for 48 hrs. |
ACKNOWLEDGEMENTS
This work was supported by grants from BioGreen 21 Program (Code20050301034420) funded by Rural Development Administration.
References
1. Balamurugan V, Kumar RM, Suryanarayana VV. Past and present vaccine development strategies for the control of foot-and-mouth disease. Acta Virol. 2004. 48:201–214.
2. Knowles NJ, Samuel AR, Davies PR, Midgley RJ, Valarcher JF. Pandemic strain of foot-and-mouth disease virus serotype O. Emerg Infect Dis. 2005. 11:1887–1893.

3. Patil PK, Suryanarayana V, Bist P, Bayry J, Natarajan C. Integrity of GH-loop of foot-and-mouth disease virus during virus inactivation: detection by epitope specific antibodies. Vaccine. 2002. 20:1163–1168.

4. Collen T, Dimarchi R, Doel TR. A T cell epitope in VP1 of foot-and-mouth disease virus is immunodominant for vaccinated cattle. J Immunol. 1991. 146:749–755.
5. Wang JH, Liang CM, Peng JM, Shieh JJ, Jong MH, Lin YL, Sieber M, Liang SM. Induction of immunity in swine by purified recombinant VP1 of foot-and-mouth disease virus. Vaccine. 2003. 21:3721–3729.

6. Sanz-Parra A, Sobrino F, Ley V. Infection with foot-and-mouth disease virus results in a rapid reduction of MHC class I surface expression. J Gen Virol. 1998. 79:433–436.

7. Yang NS, Wang JH, Lin KF, Wang CY, Kim SA, Yang YL, Jong MH, Kuo TY, Lai SS, Cheng RH, Chan MT, Liang SM. Comparative studies of the capsid precursor polypeptide P1 and the capsid protein VP1 cDNA vectors for DNA vaccination against foot-and-mouth disease virus. J Gene Med. 2005. 7:708–717.

8. Borrego B, Fernandez-Pacheco P, Ganges L, Domenech N, Fernandez-Borges N, Sobrino F, Rodríguez F. DNA vaccines expressing B and T cell epitopes can protect mice from FMDV infection in the absence of specific humoral responses. Vaccine. 2006. 24:3889–3899.

9. Collen T, Baron J, Childerstone A, Corteyn A, Doel TR, Flint M, Garcia-Valcarcel M, Parkhouse RM, Ryan MD. Heterotypic recognition of recombinant FMDV proteins by bovine T-cells: the polymerase (P3Dpol) as an immunodominant T-cell immunogen. Virus Res. 1998. 56:125–133.

10. Cedillo-Barrón L, Foster-Cuevas M, Cook A, Gutiérrez-Castañeda B, Kollnberger S, Léfevre F, Parkhouse RM. Immunogenicity of plasmids encoding T and B cell epitopes of foot-and-mouth disease virus (FMDV) in swine. Vaccine. 2003. 21:4261–4269.

11. Eriksson E, Yao F, Svensjo T, Winkler T, Slama J, Macklin MD, Andree C, McGregor M, Hinshaw V, Swain WF. In vivo gene transfer to skin and wound by microseeding. J Surg Res. 1998. 78:85–91.
12. Raz E, Carson DA, Parker SE, Parr TB, Abai AM, Aichinger G, Gromkowski SH, Singh M, Lew D, Yankauckas MA, et al. Intradermal gene immunization: the possible role of DNA uptake in the induction of cellular immunity to viruses. Proc Natl Acad Sci U S A. 1994. 91:9519–9523.

13. Bautista EM, Ferman GS, Gregg D, Brum MC, Grubman MJ, Golde WT. Constitutive expression of alpha interferon by skin dendritic cells confers resistance to infection by foot-and-mouth disease virus. J Virol. 2005. 79:4838–4847.

14. Yang JS, Kim JJ, Hwang D, Choo AY, Dang K, Maguire H, Kudchodkar S, Ramanathan MP, Weiner DB. Induction of potent Th1-type immune responses from a novel DNA vaccine for West Nile virus New York isolate (WNV-NY1999). J Infect Dis. 2001. 184:809–816.

15. Kim SA, Liang CM, Cheng IC, Cheng YC, Chiao MT, Tseng CJ, Lee F, Jong MH, Tao MH, Yang NS, Liang SM. DNA vaccination against foot-and-mouth disease via electroporation: study of molecular approaches for enhancing VP1 antigenicity. J Gene Med. 2006. 8:1182–1191.

16. Park JH, Kim SJ, Oem JK, Lee KN, Kim YJ, Kye SJ, Park JY, Joo YS. Enhanced immune response with foot and mouth disease virus VP1 and interleukin-1 fusion genes. J Vet ScI. 2006. 7:257–262.

17. Cedillo-Barrón L, Foster-Cuevas M, Belsham GJ, Lefévre F, Parkhouse RM. Induction of a protective response in swine vaccinated with DNA encoding foot-and-mouth disease virus empty capsid proteins and the 3D RNA polymerase. J Gen Virol. 2001. 82:1713–1724.

18. Zhang Q, Zhu MW, Yang YQ, Shao M, Zhang ZY, Lan HY, Yan WY, Wu JJ, Zheng ZX. A recombinant fusion protein and DNA vaccines against foot-and-mouth disease virus type Asia 1 infection in guinea pigs. Acta Virol. 2003. 47:237–243.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download