Abstract
Mitochondria are key organelles involved in energy production, functioning as the metabolic hubs of cells. Recent findings emphasize the emerging role of the mitochondrion as a key intracellular signaling platform regulating innate immune and inflammatory responses. Several mitochondrial proteins and mitochondrial reactive oxygen species have emerged as central players orchestrating the innate immune responses to pathogens and damaging ligands. This review explores our current understanding of the roles played by mitochondria in regulation of innate immunity and inflammatory responses. Recent advances in our understanding of the relationship between autophagy, mitochondria, and inflammasome activation are also briefly discussed. A comprehensive understanding of mitochondrial role in toll-like receptor-mediated innate immune responses and NLRP3 inflammasome complex activation, will facilitate development of novel therapeutics to treat various infectious, inflammatory, and autoimmune disorders.
Mitochondria are the major cellular organelle responsible for energy provision when energy needs are to be met. In addition, mitochondria have numerous quality-control mechanisms by which they protect their molecular machinery from stress and maintain cellular homeostasis (1). Earlier studies showed that infected and damaged mitochondria significantly increased their production of reactive oxygen species (ROS) and the oxidant peroxynitrite, altering the action of the electron transport chain (23). Over the last decade, it has been suggested that the inflammatory mediator such as tumor necrosis factor (TNF) is associated with mitochondrial damage by altering mitochondrial ultrastructure, inhibiting the electron transport chain, and presumably by mediating mitochondrial production of oxygen radicals (4). Recent compelling evidence suggests that mitochondria are actively involved in a multitude of cellular activities including inflammatory signaling, cell proliferation, and cell death; these pathways are attractive targets in patients with various diseases (567). Thus, inflammatory signaling and mitochondrial homeostasis may be interconnected.
Moreover, mitochondria play important roles in innate and adaptive immune responses and in inflammatory signaling (8). Mitochondrial danger signals amplify inflammatory responses (9). Importantly, metabolic remodeling orchestrated by the mitochondria is essential for appropriate control of innate and inflammatory responses (810). In this review, we will discuss the emerging functions of several mitochondrial components/proteins in terms of regulation of innate and inflammatory responses. We also present our current understanding of the functions of mitochondrial ROS and DNA in regulating innate immune and inflammatory responses. Particularly, the details of the relationship between mitophagy and inflammation have been extensively reviewed in recent articles (11) Thus, we just briefly mention current views on the functional interrelationship between mitochondrial dysfunction, inadequate autophagy, and inflammatory pathologies. Another key function of mitochondria is regulation of the immunometabolism characteristic of innate immunity. This is beyond the scope of this review and has recently been thoroughly addressed (12).
Fundamentally, the mitochondrion is the “powerhouse of the cell”, providing energy in the form of adenosine triphosphate (ATP). Mitochondria play essential roles in interconnecting diverse anabolic and catabolic processes including oxidative phosphorylation, glycolysis, the tricarboxylic acid (TCA) cycle, and fatty acid β-oxidation (713). This fundamental role of mitochondria (regulation of metabolism) involves cellular signaling networks including those controlling cell survival/death, calcium signaling, and the innate and inflammatory responses (71013). Indeed, mitochondria contain a specific genome transmitted through the female germline (14). Some mitochondrial DNA (mtDNA)-encoded proteins are structural subunits of the mitochondrial respiratory chains: including NADH dehydrogenase 1 (MTND1–MTND6 and MTND4L, complex I); cytochrome b (MTCYB, complex III); cytochrome c oxidase I (MTCO1–MTCO3, complex IV); and ATP synthase 6 (MTATP6 and MTATP8, complex V) (14). Although the electron transport chain is essential for ATP production, harmful mitochondrial ROS are generated as by-products of such transport (1516). Accumulation of mitochondrial ROS causes cell damage, inflammation, and cell death (1517).
Mitochondria are important intracellular organelle which constantly undergo dynamic process (mitochondrial dynamics) involving fission, fusion and mitochondrial autophagy (mitophagy), maintaining mitochondrial functionality and protein quality-control and maximizing the oxidative capacity in response to toxic stress (1819). Mitofusin 1/2-mediated fusion process connect two healthy mitochondria, whereas dynamin related protein 1 (Drp1) mediated fission process segregates damaged and healthy mitochondria to form robust tubular network of mitochondria, ensuring quality control via removal of damaged organelles (1819). The continuous changes in mitochondrial morphology are metabolically controlled, and are also influenced by mitophagy and macroautophagy (18). Maintenance of mitochondrial turnover is critical in terms of mitochondrial quality control and overall cellular function: damaged (potentially harmful) mitochondria that could trigger excessive inflammatory responses are cleared (20).
During invasion of a pathogen or tissue injury, the innate immune system senses and responds to a variety of pathogen- and danger-associated molecular patterns (PAMPs and DAMPs) via diverse pattern-recognition receptors (PRRs) (21). Among these, Toll-like receptors (TLRs) are the most widely studied: these receptors trigger complicated intracellular signaling cascades and activate host defenses (2122). The extracellular leucine-rich repeat (LRR) domains of TLRs recognize various ligands of bacteria, viruses, fungi, and protozoa. To date, 10 functional TLRs (TLR1 to TLR10) have been identified in humans (23). Mouse TLR11 recognizes protozoan profilin-like proteins and uropathogenic bacteria, but is non-functional in humans (2425). Unlike TLRs, NOD-like receptors (NLRs) sense PAMPs and DAMPs in the intracellular cytosolic compartment (26). Many NLRs have been identified: these include Nod1, Nod2, NLRP3, NLRC4, NLRP6, NLRX1, NLRC3, NLRC5, and NLRP4 (27). Upon binding of PAMPs and DAMPs to the innate immune receptors, intracellular signaling cascades are activated via recruitment of adaptor proteins and cellular kinases, culminating in activation of the nuclear factor (NF)-κB and mitogen-activated protein kinase (MAPK) pathways (2728).
In addition, several types of NLRs and AIM2-like receptors (ALRs) including NLRP3 (NOD-, LRR- and pyrin domain-containing [protein] 3), NLRP1, NLRP6, NLRP7, NLRC4, and AIM2 can form inflammasomes (large protein complexes) regulating interleukin-1β (IL-1β) and IL-18 secretion (272930). Activation of the NLRP3 inflammasome complex usually requires a two-step signal (priming and activation). Upregulated TLR signaling activates the NF-κB pathway and induces transcription of pro-IL-1β and NLRP3, thereby serving to prime activation of the inflammasome complex (31). Although the second signal induced by a variety of PAMPs and DAMPs is not fully characterized, that signal triggers oligomerization of inflammasome components and caspase-1 auto-activation, followed by assembly of the NLRP3 inflammasome (32). Importantly, activation of NLRP3 and AIM2 induces nucleation of PYD filaments and clustering of the CARD proteins of the ASC adaptor, completing inflammasome assembly (33).
Upon viral infection, retinoic acid-inducible gene 1 (RIG-I)-like receptors (RLRs) RIG-I, MDA5, and LGP2 discriminate between host RNA (self RNA) and viral RNA (non-self RNA) and specifically sense viral RNA produced by both RNA and DNA viruses at the cytoplasm (3435). Upon RLR signaling, RIG-I and MDA5 specifically ubiquitinated by TRIM25 and TRIM65 respectively (3637) bind to the common adaptor MAVS/Cardif/IPS-1/VISA by CARD-CARD interaction leading to activation of downstream IKK family (IKKɛ, TBK1, and IKKα/β/γ). These kinases next cause activation of downstream NF-κB signaling pathway and also induce phosphorylation and subsequent homodimerization of IFN-regulatory factors 3 and 7 (IRF3/7) (transcription factors) respectively, thereby resulting in nuclear translocation of dimerized phosphorylated IRF3 and IRF7, in turn promoting type-I interferon (IFN-I) synthesis which, finally, transcriptionally activates many IFN-stimulated genes (ISGs) (3538). RIG-I and MDA5 signaling pathways are summarized in Fig. 1. In vivo, the actions of the TLR, NLR, and RLR signaling systems are spatially and temporally co-ordinated to allow the generation of appropriate and concerted responses to effectors via integration of many immune signal transductions (39). The magnitude of an immune reaction must be tightly regulated to avoid immunopathologies (39). Uncontrolled activation of innate responses is strongly associated with the pathologies of various inflammatory and autoimmune diseases (40).
Mitochondrial antiviral signaling (MAVS) protein, an outer mitochondrial membrane protein, is the key mediator of the innate immune response upon viral infections (4142). Earlier studies showed that the MAVS protein was a pivotal signaling adaptor, inducing antiviral and inflammatory pathways via activation of NF-κB and IRF-3 during development of innate immune responses to RNA viruses (4243). Such effects seem to be specific to viral infection. MAVS protein was not essential for induction of interferon production in response to cytosolic DNA or intracellular listerial infection (44). Further studies showed that RIG-I signaling triggered formation of large prion-like MAVS protein aggregates on the mitochondrial membrane, activating IRF3 responses (45). MAVS protein was also required for dsDNA-induced IFN-β transcriptional activation in a human hepatoma cell line (Huh-7) (46). Moreover, MAVS protein was essential for induction of type I IFN and the antiviral response to respiratory syncytial virus. MAVS protein acted in concert with the TLR adaptor MyD88 (47). Previous studies also showed that MAVS protein was required for the antibacterial responses of endothelial cells to Chlamydophila pneumoniae infection (48). Interestingly, MAVS protein is essential for maintenance of intestinal homeostasis, presumably because the protein monitors intestinal commensal bacteria. MAVS protein deficiency increased both the severity of the response to colitis and mortality (49). Also, RLR activation enhanced mitochondrial elongation and fusion, promoting MAVS protein-mediated signaling (50). In addition, MAVS protein played critical roles in activation of the NLRP inflammasome, and subsequent IL-1b production, by mediating NLRP3 recruitment to mitochondria (51). Apart from regulating antiviral type I IFN responses, the MAVS protein also triggered dsRNA-induced apoptosis by interacting with caspase-8; the Bax/Bak pathway was not involved (52). Via MAVS-MAPK kinase 7 (MKK7)-JNK2 signaling, MAVS protein was involved in the regulation of Sendai virus-induced apoptosis, and the host defense to viral infection (53). Importantly, the MAVS protein-dependent type I IFN response was important in controlling Plasmodium replication in the liver (54).
Emerging evidence allows us to begin to understand the molecular mechanisms by which the MAVS signalosome is controlled to regulate antiviral responses. It was earlier shown that a TRAF-interaction motif (TIM) within the MAVS protein interacted directly with the TRAF domain of TRAF3 to activate the antiviral immune response (55). Recent studies have shown that the tripartite motif 14 (TRIM14) interacts with MAVS protein and NF-κB within the MAVS signalosome, thereby mediating the immune response during viral infection (56). It was also shown that transcription factor ELF4, induced by type I IFN, positively regulated IFN production via interaction with and activation of the MAVS-TBK1 complex, enhancing the response to West Nile virus in mice (57). In addition, the tyrosine kinase c-Abl positively regulated MAVS protein function via physical and functional interaction (58). Silencing of c-Abl inhibited the MAVS protein-mediated innate immune response via regulation of NF-κB and IRF3 signaling (58). Indeed, several TRAF proteins, including TRAF2, TRAF5, and TRAF6, were recruited to MAVS protein polymers, activating IRF3 signaling and the antiviral immune response (59). Although the precise mechanism remains unclear, it was recently shown that the enzyme, pyruvate carboxylase (PC), is essential for virus-triggered activation of the innate immune response; the enzyme targets the MAVS signalosome (60).
On the other hand, poly(RC)-binding protein 2 (PCBP2) negatively regulated MAVS protein-mediated antiviral signaling (61). The same authors also showed that PCBP1 (which is functionally similar to PCBP2) inhibited the MAVS protein-mediated antiviral immune response by triggering MAVS protein degradation via Lys48-linked polyubiquitination (62). Notably, PCBP1 and PCBP2 synergistically inhibited MAVS protein signaling (62). In addition, the proteasome PSMA7 (alpha 4) subunit, which interacts with the MAVS protein, negatively regulated the RIG-1- and MAVS protein-mediated type I IFN responses and antiviral activities (63). Further study showed that NLRX1 interfered with the interaction between the MAVS protein and RIG-I, finely tuning type I IFN signaling and the cytokine response (64). Reports on how NLRX1 regulates MAVS protein signaling are conflicting. However, NLRX1-deficient mice did not exhibit alterations in their antiviral or inflammatory responses, as compared to control mice (65).
More recently, the cytochrome c oxidase (CcO) complex subunit, COX5B, a component of the mitochondrial electron transport system, was shown to be physically associated with the MAVS protein and to inhibit MAVS proteinmediated antiviral immunity via autophagy- and ROS-dependent pathways (66). The UBX-domain-containing protein UBXN1 inhibited RNA virus-mediated antiviral signaling by binding to the MAVS protein, preventing oligomerization thereof (67). Further study showed that the Smad ubiquitin regulatory factor (Smurf) 2 negatively regulated antiviral type I IFN responses by interacting with the MAVS protein, triggering proteasome-mediated degradation (68). Recent studies have revealed the autoinhibitory mechanisms by which MAVS protein-mediated antiviral activity is tightly regulated even under unstimulated conditions, to prevent spontaneous RIG-I activation (69). Very recently, the NS3 protein of dengue virus has been shown to bind to 14-3-3ɛ, an essential cellular protein mediating the cytosol-to-mitochondrial membrane translocation of RIG-I, thereby preventing translocation of RIG-I to the MAVS protein, which would inhibit antiviral immunity (70).
Another recent study reported a novel function of the insulin receptor tyrosine kinase substrate (IRTKS), which plays crucial roles in actin bundling and insulin signaling. IRTKS negatively regulated MAVS protein signaling by sumoylation of PCBP2, which then interacted with MAVS protein to trigger degradation thereof (71). Moreover, the mitochondrion-resident E3 ligase, MARCH5, inhibited MAVS protein-mediated antiviral immune responses and excessive inflammatory reactions by binding to the protein, promoting proteasome-mediated degradation (72). In addition, a recent study has shown that protein phosphatase magnesium-dependent 1A (PPM1A; also known as PP2Cα) acts on the MAVS protein, inhibiting RLR-MAVS protein signaling by targeting and dephosphorylating both MAVS protein and TBK1/IKKɛ (73). Another MAVS protein-interacting protein, the adaptor TAX1BP1, negatively regulated apoptosis triggered by infection with RNA viruses including vesicular stomatitis virus and Sendai virus (74). TAX1BP1 recruited the E3 ligase Itch to the MAVS protein, triggering ubiquitination and proteasomal degradation (74). In summary, many positive and negative regulators co-ordinate the antiviral immune responses by interacting with and post-translationally modifying the MAVS protein of the mitochondrion (Fig. 2).
Several mitochondrial proteins involved in controlling mitochondrial morphology are also known to play roles in the fine-tuning of innate immune responses (75). Earlier studies showed that the mitochondrial adaptor protein, ECSIT (evolutionarily conserved signaling intermediate in Toll pathways), interacted with tumor necrosis factor receptor-associated factor 6 (TRAF6), resulting in upregulation of mitochondrial ROS production in macrophages, which is essential for bactericidal activity (76). A recent study showed that kinases Mst1 and Mst2 promoted TLR-induced assembly of the TRAF6-ECSIT complex via Rac activation, thus triggering recruitment of mitochondria to phagosomes and enhancing bactericidal activity (77). Indeed, the ECSIT-TAK1-TRAF6 complex was essential for activation of TLR4-induced NF-κB signaling and cytokine production in monocytic cells (78). Another study found that ECSIT became associated with MAVS protein on the mitochondrial surface, mediating bridging of the MAVS protein to RIG-I or MDA5, in turn inducing activation of the antiviral response via upregulation of IFN-regulatory factor 3 (IRF3) and increased expression of IFNB1 during viral infection (79).
The mitochondrial outer membrane protein voltage-dependent anion channel 1 (VDAC1) protein is essential for mitochondrial ROS production and NLRP3 inflammasome activation (80). Previous studies showed that MARCH5 (a mitochondrial outer membrane protein) and E3 ligase activated TLR7 signaling via an interaction with TANK. The interaction involved catalysis of K63-linked TANK poly-ubiquitination on lysines 229, 233, 280, 302, and 306 (75). As mentioned above, MARCH5 serves as a negative regulator of MAVS protein activity by interacting with and ubiquitinating the MAVS protein, promoting proteasome-mediated degradation, thus inhibiting excessive immune responses (72). The ubiquitin ligase PARKIN, which is essential for mitochondrial protein ubiquitination, was also found to be essential for ubiquitin-mediated autophagy and LUBAC-mediated MAVS ubiquitination followed by attenuation in downstream IFN signaling in HBV infected cells (81), and mounting of a host defense against Mycobacterium tuberculosis (82).
In contrast, several mitochondrial proteins have been shown to negatively regulate inflammasome activation. Previous work explored the regulatory role played by the anti-apoptotic Bcl-2 protein during such activation. Both Bcl-2 transgenic macrophages, and macrophages in which Bcl-2 was overexpressed, exhibited significantly reduced IL-1β production in response to NLRP3-mediated stimuli, suggesting that inhibition of apoptosis affected IL-1β maturation and secretion triggered by NLRP3 activation (83). Also, the anti-apoptotic proteins, Bcl-2 and Bcl-X(L), suppressed activation of the NALP1 inflammasome (84). Indeed, mitochondria contain a signaling platform, the mitoxosome, which integrates the multiple signaling pathways associated with viral recognition and cellular stress to co-ordinate the antiviral response; the mitoxosome is the point of convergence of the relevant pathways (20).
Mitochondrial ROS have been suggested to be key signaling activators of the innate immune responses triggered by TLR agonists. Earlier studies found that lipopolysaccharide (LPS)-induced synthesis of proinflammatory cytokines (including TNF and IL-6) was modulated by scavenging of mitochondrial ROS (85). Enhancement of the innate immune response in patients with TNF receptorassociated periodic syndrome (TRAPS), an autoinflammatory disorder caused by missense mutations in the type 1 TNF receptor (TNFR1), is at least partially attributable to increased mitochondrial ROS generation (85). Previous studies showed that both mitochondrial- and Duox2-generated ROS were essential for reduction of influenza A virus titers, and induction of antiviral innate immune responses, via regulation of IFN-λ secretion by normal human nasal epithelial cells (86). We recently found that upregulation of TLR4-induced proinflammatory cytokine production in small, heterodimer partner-deficient macrophages was mediated via mitochondrial ROS (87). In microglia, LPS-induced inflammatory signaling and cytokine production are mediated by mitochondrial ROS (88).
As mentioned above, the kinases Mst1 and Mst2, the closest mammalian homologs of the Drosophila kinase Hippo, are critically involved in the optimal generation of mitochondrial ROS in phagocytes responding to TLR signaling, thus regulating antimicrobial responses (77). A very recent study found that increased mitochondrial ROS generation after succinate oxidation triggered IL-1β synthesis attributable to HIF-1α stabilization (89). Previously, mtDNA was shown to be spared during autophagy, followed by induction of TLR9-mediated inflammatory responses in cardiomyocytes, in turn triggering myocarditis and dilated cardiomyopathy (90). After injury or septic shock, mitochondrial DAMPs (including mtDNA and peptides) are released into the circulation, causing cellular injury and pathological endothelial permeability (91). mtDNA has recently been shown to exert an antiviral function. MtDNA released to the cytosol is recognized by the DNA sensor cGAS (also termed MB21D1) and activates STING (also termed TMEM173)-IRF3 signaling to enhance the expression of interferon-stimulated genes and the type I interferon responses (92).
Many studies have defined the critical roles played by mitochondria in inflammasome activation. Notably, mitochondrial ROS play a critical role in activation of the NLRP3 inflammasome complex (7). Stimuli of the NLRP3 inflammasome include ATP, alum, nigericin, and Chlamydia pneumoniae; all irreversibly reduce the mitochondrial membrane potential and oxygen consumption rate (OCR), but increase mitochondrial ROS generation (83). mtDNA released into the cytosol by apoptotic signaling during apoptosis bound to NLRP3 and activated the NLRP3 inflammasome (83). Interestingly, the AIM2 inflammasome was activated by exogenous, but not endogenous, mtDNA, suggesting that cytosolic translocation of mtDNA activates the NLRP3 inflammasome in a specific manner (8393). In addition, rotenone-induced impairment of the mitochondrial electron transport chain and mitochondrial hyperpolarization constitute priming signals for caspase-1 processing and NLRP3 inflammasome activation only in the presence of ATP (94).
Recent studies have shown that inducers of NLRP3 inflammasome activation alter mitochondrial homeostasis and reduce the concentration of the coenzyme NAD(+), triggering accumulation of acetylated α-tubulin and dynein-dependent mitochondrial transport (95). The cited author argued that microtubule-dependent transport of mitochondria to NLRP3 on the endoplasmic reticulum was essential for assembly of the NLRP3 inflammasome complex (95). Secondary NLRP3 signals, including ATP, also induce the release of oxidized mtDNA, which can bind to the NLRP3 inflammasome and directly induce activation (83). In macrophages, both LPS and ATP (NLRP3 inflammasome stimuli) significantly increased the cytosolic accumulation of mtDNA, which acted as a co-activator of caspase-1 (93). No cytosolic translocation of mtDNA was observed in NLRP3- or ASC-knockout macrophages, suggesting that inflammasome activation per se mediates release of mtDNA into the cytosol (93). In addition, macrophages of the mitochondrial DNA-depleted Rho 0 (ρ0) phenotype exhibited significant attenuation of caspase-1 activation and IL-1β cleavage, but did not contain reduced levels of pro-IL-1β (8393). Thus, mitochondrial ROS production and DNA translocation into the cytosol play key roles in the innate regulation and control of inflammatory responses.
Autophagy is a lysosome-mediated intracellular degradation pathway allowing removal of damaged organelles. The selective or non-selective autophagic removal of impaired mitochondria (selective or non-selective mitophagy) controls mitochondrial quality which is indispensable to sustain cell homeostasis. This mitophagic process may affect the formation of inflammasome followed by activation of inflammatory responses (96). Growing evidences suggest that defective or incomplete mitophagic process may lead to aberrant activation of inflammatory response (1197). In the present review, we will only briefly discuss recent progress regarding autophagic/mitophagic modulation of various functions of mitochondria associated with regulation of innate immune response.
Selective or non-selective mitophagic process is completed by lysosomal delivery of autophagosome engulfing impaired mitochondria (98). Several evidences have shown that incomplete autophagic clearance of damaged mitochondria may trigger aberrant inflammasome activation and lead to a variety of human inflammatory diseases (1198). In macrophages, autophagy blockade increases the production of mitochondrial ROS which induces mitochondrial damage, in turn activating the inflammasome (8099). In LC3- or Beclin1-deficient cells, even the basal mtROS levels were elevated, and cytosolic leakage of mtDNA induces activation of the NLRP3 inflammasome and subsequent increase of IL-1β secretion (93).
During RLR signaling, autophagy-defective cells accumulate damaged mitochondria, triggering amplification of inflammatory signaling (20). In this condition, a couple of suppositions might be considered: 1) Increase of MAVS concomitant with accumulation of damaged mitochondria in autophagy deficient MEFs and macrophages; 2) Maintenance of high levels of mtROS concomitant with accumulation of damaged mitochondria, in turn activating inflammatory signaling (20100).
Recent studies have shown that autophagy is essential for the clearance of damaged mitochondria, and that scavenging of mitochondria-associated ROS increased the survival of virus-specific natural killer cells (101). The mitochondrial proteins BCL2/adenovirus E1B 19-kDa interacting protein 3 (BNIP3) and BNIP3-like (BNIP3L, NIX) play critical roles in this form of this protective mitophagy. The pro-survival signals facilitate formation of memory NK cells (101). Another study showed that initiation of autophagy by FoxO1 mediates NK cell development and effective antiviral functionality (102). Together, these data strongly suggest that activation of autophagy, which is essential to ensure mitochondrial homeostasis, is also critical in terms of the fine-tuning and appropriate induction of innate immune activation.
It is becoming apparent that mitochondria are crucial organelles, not only because of their essential role in energy production but also because they co-ordinate the signaling networks associated with innate immunity, autophagy, and inflammation. We have focused on the essential roles played by mitochondrial proteins in regulating innate immunity and inflammatory responses. Of the various mitochondrial components, the MAVS protein is key in terms of regulation of RLR signaling and antiviral immune responses. Many positive and negative regulators of the MAVS protein have been identified; these act as fine modulators of innate antiviral immune responses. Other mitochondrial proteins, including VDAC, ECSIT, and MARCH5, have been suggested to be associated with the immune and inflammatory responses. Identification of further relevant proteins, coupled with an understanding of how they regulate innate immune responses, will increase the number of useful targets for therapies aimed at treating infectious and inflammatory diseases. In addition, mitochondria control many innate signaling pathways via ROS and mtDNA. Emerging evidence suggests that both autophagy and mitophagy play crucial roles in the control of mitochondrial homeostasis and regulation of innate and inflammatory responses. An intricate interplay is evident between mitochondria, autophagy, and the inflammatory response. A fuller understanding of this process will lead to the definition of new therapeutic strategies for acute and chronic pathological and inflammatory disorders.
Figures and Tables
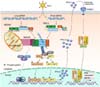 | Figure 1Overview of RLRs signaling pathway. Retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs) recognize the genomic RNA or RNA replication intermediates of viruses as cytoplasmic RNA sensors. Following viral infection, melanoma differentiation-associated protein 5 (MDA5) recognizes cytoplasmic viral long-scale double-stranded RNA (dsRNA) whereas RIG-I recognizes short viral dsRNA (non-self RNA). Upon recognition of viral dsRNA, MDA5 and RIG-I specifically ubiquitinated by TRIM65 and TRIM25 respectively initiate antiviral innate immune response via specific interaction with mitochondrial antiviral signaling protein (MAVS) by CARD-CARD interaction. MAVS modulates nuclear factor-kB (NF-kB) activity via IKK complex (IKK α/β/γ) activation. MAVS also interacts with TRAFs translocated onto mitochondria upon viral infection and subsequently induces recruitment of TBK1 and IκB kinase-ɛ (IKKɛ) to promote phosphorylation of interferon (IFN) regulatory factor 3 (IRF3) and IRF7. Phosphorylated IRF3 and IRF7 cause their homo-dimerization which is translocated to the nucleus. In the nucleus, homo-dimerized IRF3 and IRF7 bind to specific binding sites in the IFNβ and IFNα promoter respectively to stimulate type I IFN synthesis. Secreted type I IFNs (IFNβ and IFNα) binds to interferon alpha and beta receptor subunit 1 (IFNAR1) and subsequently induces phosphorylation of signal transducer and activator of transcription 1 (STAT1) and STAT2, leading to the induction of nuclear translocation of IRF7/STAT1/STAT2 complex followed by promotion of IFN-stimulated genes (ISGs) transcription. Solid arrows indicate direct signaling. Dashed arrows indicate indirect signaling. |
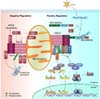 | Figure 2Positive and negative regulation of MAVS-mediated antiviral signaling pathway. Upon viral infection, sensing of viral dsRNA by RLRs induces the formation of MAVS signalosome on mitochondria followed by promotion in downstream IFN synthesis. TNF receptor-associated factor 6 (TRAF6), TNFR1–associated death domain protein (TRADD), tripartite motif 14 (TRIM14) and pyruvate carboxylase (PC) modulates canonical NF-κB signaling pathway. Activated IκB kinase (IKK) complex (IKK α/β/γ) induces phosphorylation of NF–κB inhibitor–α (IκBα), resulting in NF–κB nuclear translocation and transcriptional activation of proinflammatory cytokines gene expression. MAVS also interacts with TRAF2/3, TANK, IKKe and TBK1. TBK1-mediated phosphorylation of IRF3 and IRF7 and subsequent their dimerization promotes type I IFN gene expression through nuclear translocation. Various molecules are involved in negative regulation of MAVS signaling. Poly(RC)-binding protein (PCBP) 1 and PCBP2 induces Lys48-linked polyubiquitination of MAVS, resulting in its proteasomal proteosomal degradation. Also, Smad ubiquitin regulatory factor 2 (Smurf2) binding to MAVS reduces antiviral type I IFN production through proteosomal degradation of MAVS. 20S proteasomal subunit PSMA7 negatively regulates MAVS signaling by promoting degradation, NLR family member X1 (NLRX1) downregulates type I IFN production by inhibiting between MAVS and RIG-I direct interaction. Cytochrome c oxidase (CcO) complex subunit (COX5B) downregulates type I IFN production by physical interaction with MAVS. UBX-domain-containing protein UBXN1 inhibit MAVS oligomerization, resulting in inhibition of antiviral signaling pathway. |
ACKNOWLEDGEMENTS
We are indebted to current and past members of our laboratory for discussions and investigations that contributed to this article. This work was supported by the research fund of Chungnam National University and the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (No. NRF-2015M3C9A2054326). I apologize to colleagues whose work and publications could not be referenced owing to space constraints. The authors have no financial conflict of interests.
References
1. Cheng Z, Ristow M. Mitochondria and metabolic homeostasis. Antioxid Redox Signal. 2013; 19:240–242.

2. Taylor DE, Ghio AJ, Piantadosi CA. Reactive oxygen species produced by liver mitochondria of rats in sepsis. Arch Biochem Biophys. 1995; 316:70–76.

3. Kurose I, Miura S, Fukumura D, Yonei Y, Saito H, Tada S, Suematsu M, Tsuchiya M. Nitric oxide mediates Kupffer cell-induced reduction of mitochondrial energization in hepatoma cells: a comparison with oxidative burst. Cancer Res. 1993; 53:2676–2682.
4. Schulze-Osthoff K, Bakker AC, Vanhaesebroeck B, Beyaert R, Jacob WA, Fiers W. Cytotoxic activity of tumor necrosis factor is mediated by early damage of mitochondrial functions. Evidence for the involvement of mitochondrial radical generation. J Biol Chem. 1992; 267:5317–5323.

5. Cloonan SM, Choi AM. Mitochondria: sensors and mediators of innate immune receptor signaling. Curr Opin Microbiol. 2013; 16:327–338.

6. Dromparis P, Michelakis ED. Mitochondria in vascular health and disease. Annu Rev Physiol. 2013; 75:95–126.

7. Gurung P, Lukens JR, Kanneganti TD. Mitochondria: diversity in the regulation of the NLRP3 inflammasome. Trends Mol Med. 2015; 21:193–201.

8. Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015; 42:406–417.

9. Lopez-Armada MJ, Riveiro-Naveira RR, Vaamonde-Garcia C, Valcarcel-Ares MN. Mitochondrial dysfunction and the inflammatory response. Mitochondrion. 2013; 13:106–118.

10. Sandhir R, Halder A, Sunkaria A. Mitochondria as a centrally positioned hub in the innate immune response. Biochim Biophys Acta. 2016; DOI: 10.1016/j.bbadis.2016.10.020.

11. Kim MJ, Yoon JH, Ryu JH. Mitophagy: a balance regulator of NLRP3 inflammasome activation. BMB Rep. 2016; 49:529–535.

12. Kugelberg E. Immunometabolism: Mitochondria adapt to bacteria. Nat Rev Immunol. 2016; 16:464–465.
13. Wasilewski M, Chojnacka K, Chacinska A. Protein trafficking at the crossroads to mitochondria. Biochim Biophys Acta. 2017; 1864:125–137.

14. Stewart JB, Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet. 2015; 16:530–542.

15. Hekimi S, Wang Y, Noe A. Mitochondrial ROS and the effectors of the intrinsic apoptotic pathway in aging cells: The discerning killers! Front Genet. 2016; 7:161.

16. Dan Dunn J, Alvarez LA, Zhang X, Soldati T. Reactive oxygen species and mitochondria: A nexus of cellular homeostasis. Redox Biol. 2015; 6:472–485.

17. Rimessi A, Previati M, Nigro F, Wieckowski MR, Pinton P. Mitochondrial reactive oxygen species and inflammation: Molecular mechanisms, diseases and promising therapies. Int J Biochem Cell Biol. 2016; 81:281–293.

18. Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012; 337:1062–1065.

19. Haroon S, Vermulst M. Linking mitochondrial dynamics to mitochondrial protein quality control. Curr Opin Genet Dev. 2016; 38:68–74.

20. Tal MC, Iwasaki A. Mitoxosome: a mitochondrial platform for cross-talk between cellular stress and antiviral signaling. Immunol Rev. 2011; 243:215–234.

21. Brubaker SW, Iwasaki A. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol. 2015; 33:257–290.

22. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010; 11:373–384.

23. Skevaki C, Pararas M, Kostelidou K, Tsakris A, Routsias JG. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious diseases. Clin Exp Immunol. 2015; 180:165–177.

24. Yarovinsky F, Zhang D, Andersen JF, Bannenberg GL, Serhan CN, Hayden MS, Hieny S, Sutterwala FS, Flavell RA, Ghosh S, Sher A. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science. 2005; 308:1626–1629.

25. Zhang D, Zhang G, Hayden MS, Greenblatt MB, Bussey C, Flavell RA, Ghosh S. A toll-like receptor that prevents infection by uropathogenic bacteria. Science. 2004; 303:1522–1526.

26. Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006; 442:39–44.

27. Elinav E, Strowig T, Henao-Mejia J, Flavell RA. Regulation of the antimicrobial response by NLR proteins. Immunity. 2011; 34:665–679.

29. Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012; 481:278–286.

30. Franchi L, Munoz-Planillo R, Nunez G. Sensing and reacting to microbes through the inflammasomes. Nat Immunol. 2012; 13:325–332.

31. Tall AR, Yvan-Charvet L. Cholesterol, inflammation and innate immunity. Nat Rev Immunol. 2015; 15:104–116.

32. Gombault A, Baron L, Couillin I. ATP release and purinergic signaling in NLRP3 inflammasome activation. Front Immunol. 2012; 3:414.

33. Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, Egelman EH. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014; 156:1193–1206.

34. Rodriguez KR, Bruns AM, Horvath CM. MDA5 and LGP2: accomplices and antagonists of antiviral signal transduction. J Virol. 2014; 88:8194–8200.

35. Chan YK, Gack MU. RIG-I-like receptor regulation in virus infection and immunity. Curr Opin Virol. 2015; 12:7–14.

36. Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007; 446:916–920.

37. Lang X, Tang T, Jin T, Ding C, Zhou R, Jiang W. TRIM65-catalized ubiquitination is essential for MDA5-mediated antiviral innate immunity. J Exp Med. 2017; 214:459–473.

38. Eisenacher K, Krug A. Regulation of RLR-mediated innate immune signaling--it is all about keeping the balance. Eur J Cell Biol. 2012; 91:36–47.
39. Thaiss CA, Levy M, Itav S, Elinav E. Integration of Innate Immune Signaling. Trends Immunol. 2016; 37:84–101.

40. Li M, Zhou Y, Feng G, Su SB. The critical role of Toll-like receptor signaling pathways in the induction and progression of autoimmune diseases. Curr Mol Med. 2009; 9:365–374.

41. Vazquez C, Horner SM. MAVS coordination of antiviral innate immunity. J Virol. 2015; 89:6974–6977.

42. Belgnaoui SM, Paz S, Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol. 2011; 23:564–572.

43. Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005; 122:669–682.

44. Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006; 24:633–642.

45. Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011; 146:448–461.

46. Cheng G, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. Double-stranded DNA and double-stranded RNA induce a common antiviral signaling pathway in human cells. Proc Natl Acad Sci U S A. 2007; 104:9035–9040.

47. Bhoj VG, Sun Q, Bhoj EJ, Somers C, Chen X, Torres JP, Mejias A, Gomez AM, Jafri H, Ramilo O, Chen ZJ. MAVS and MyD88 are essential for innate immunity but not cytotoxic T lymphocyte response against respiratory syncytial virus. Proc Natl Acad Sci U S A. 2008; 105:14046–14051.

48. Buss C, Opitz B, Hocke AC, Lippmann J, van Laak V, Hippenstiel S, Krüll M, Suttorp N, Eitel J. Essential role of mitochondrial antiviral signaling, IFN regulatory factor (IRF)3, and IRF7 in Chlamydophila pneumoniae-mediated IFN-beta response and control of bacterial replication in human endothelial cells. J Immunol. 2010; 184:3072–3078.
49. Li XD, Chiu YH, Ismail AS, Behrendt CL, Wight-Carter M, Hooper LV, Chen ZJ. Mitochondrial antiviral signaling protein (MAVS) monitors commensal bacteria and induces an immune response that prevents experimental colitis. Proc Natl Acad Sci U S A. 2011; 108:17390–17395.

50. Castanier C, Chiu YH, Ismail AS, Behrendt CL, Wight-Carter M, Hooper LV, Chen ZJ. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 2010; 11:133–138.

51. Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 2013; 153:348–361.

52. El Maadidi S, Faletti L, Berg B, Wenzl C, Wieland K, Chen ZJ, Maurer U, Borner C. A novel mitochondrial MAVS/Caspase-8 platform links RNA virus-induced innate antiviral signaling to Bax/Bak-independent apoptosis. J Immunol. 2014; 192:1171–1183.

53. Huang Y, Liu H, Li S, Tang Y, Wei B, Yu H, Wang C. MAVS-MKK7-JNK2 defines a novel apoptotic signaling pathway during viral infection. PLoS Pathog. 2014; 10:e1004020.

54. Liehl P, Zuzarte-Luis V, Chan J, Zillinger T, Baptista F, Carapau D, Konert M, Hanson KK, Carret C, Lassnig C, Müller M, Kalinke U, Saeed M, Chora AF, Golenbock DT, Strobl B, Prudencio M, Coelho LP, Kappe SH, Superti-Furga G, Pichlmair A, Vigário AM, Rice CM, Fitzgerald KA, Barchet W, Mota MM. Host-cell sensors for Plasmodium activate innate immunity against liver-stage infection. Nat Med. 2014; 20:47–53.

55. Saha SK, Pietras EM, He JQ, Kang JR, Liu SY, Oganesyan G, Shahangian A, Zarnegar B, Shiba TL, Wang Y, Cheng G. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 2006; 25:3257–3263.

56. Zhou Z, Jia X, Xue Q, Dou Z, Ma Y, Zhao Z, Jiang Z, He B, Jin Q, Wang J. TRIM14 is a mitochondrial adaptor that facilitates retinoic acid-inducible gene-I-like receptor-mediated innate immune response. Proc Natl Acad Sci U S A. 2014; 111:E245–E254.

57. You F, Wang P, Yang L, Yang G, Zhao YO, Qian F, Walker W, Sutton R, Montgomery R, Lin R, Iwasaki A, Fikrig E. ELF4 is critical for induction of type I interferon and the host antiviral response. Nat Immunol. 2013; 14:1237–1246.

58. Song T, Wei C, Zheng Z, Xu Y, Cheng X, Yuan Y, Guan K, Zhang Y, Ma Q, Shi W, Zhong H. c-Abl tyrosine kinase interacts with MAVS and regulates innate immune response. FEBS Lett. 2010; 584:33–38.

59. Liu S, Wei C, Zheng Z, Xu Y, Cheng X, Yuan Y, Guan K, Zhang Y, Ma Q, Shi W, Zhong H. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife. 2013; 2:e00785.

60. Cao Z, Zhou Y, Zhu S, Feng J, Chen X, Liu S, Peng N, Yang X, Xu G, Zhu Y. Pyruvate Carboxylase Activates the RIG-I-like Receptor-Mediated Antiviral Immune Response by Targeting the MAVS signalosome. Sci Rep. 2016; 6:22002.

61. You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, Jiang Z. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol. 2009; 10:1300–1308.

62. Zhou X, You F, Chen H, Jiang Z. Poly(C)-binding protein 1 (PCBP1) mediates housekeeping degradation of mitochondrial antiviral signaling (MAVS). Cell Res. 2012; 22:717–727.

63. Jia Y, Song T, Wei C, Ni C, Zheng Z, Xu Q, Ma H, Li L, Zhang Y, He X, Xu Y, Shi W, Zhong H. Negative regulation of MAVS-mediated innate immune response by PSMA7. J Immunol. 2009; 183:4241–4248.

64. Allen IC, Song T, Wei C, Ni C, Zheng Z, Xu Q, Ma H, Li L, Zhang Y, He X, Xu Y, Shi W, Zhong H. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity. 2011; 34:854–865.

65. Soares F, Tattoli I, Wortzman ME, Arnoult D, Philpott DJ, Girardin SE. NLRX1 does not inhibit MAVS-dependent antiviral signalling. Innate Immun. 2013; 19:438–448.

66. Zhao Y, Sun X, Nie X, Sun L, Tang TS, Chen D, Sun Q. COX5B regulates MAVS-mediated antiviral signaling through interaction with ATG5 and repressing ROS production. PLoS Pathog. 2012; 8:e1003086.

67. Wang P, Yang L, Cheng G, Yang G, Xu Z, You F, Sun Q, Lin R, Fikrig E, Sutton RE. UBXN1 interferes with Rig-I-like receptor-mediated antiviral immune response by targeting MAVS. Cell Rep. 2013; 3:1057–1070.

68. Pan Y, Li R, Meng JL, Mao HT, Zhang Y, Zhang J. Smurf2 negatively modulates RIG-I-dependent antiviral response by targeting VISA/MAVS for ubiquitination and degradation. J Immunol. 2014; 192:4758–4764.

69. Shi Y, Yuan B, Qi N, Zhu W, Su J, Li X, Qi P, Zhang D, Hou F. An autoinhibitory mechanism modulates MAVS activity in antiviral innate immune response. Nat Commun. 2015; 6:7811.

70. Chan YK, Gack MU. A phosphomimetic-based mechanism of dengue virus to antagonize innate immunity. Nat Immunol. 2016; 17:523–530.

71. Xia P, Wang S, Xiong Z, Ye B, Huang LY, Han ZG, Fan Z. IRTKS negatively regulates antiviral immunity through PCBP2 sumoylation-mediated MAVS degradation. Nat Commun. 2015; 6:8132.

72. Yoo YS, Park YY, Kim JH, Cho H, Kim SH, Lee HS, Kim TH, Kim YS, Lee Y, Kim CJ, Jung JU, Lee JS, Cho H. The mitochondrial ubiquitin ligase MARCH5 resolves MAVS aggregates during antiviral signalling. Nat Commun. 2015; 6:7910.

73. Xiang W, Zhang Q, Lin X, Wu S, Zhou Y, Meng F, Fan Y, Shen T, Xiao M, Xia Z, Zou J, Feng XH, Xu P. PPM1A silences cytosolic RNA sensing and antiviral defense through direct dephosphorylation of MAVS and TBK1. Sci Adv. 2016; 2:e1501889.

74. Choi YB, Shembade N, Parvatiyar K, Balachandran S, Harhaj EW. TAX1BP1 Restrains Virus-Induced Apoptosis by Facilitating Itch-Mediated Degradation of the Mitochondrial Adaptor MAVS. Mol Cell Biol. 2017; 37:pii: e00422-16.

75. Shi HX, Liu X, Wang Q, Tang PP, Liu XY, Shan YF, Wang C. Mitochondrial ubiquitin ligase MARCH5 promotes TLR7 signaling by attenuating TANK action. PLoS Pathog. 2011; 7:e1002057.

76. West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, Walsh MC, Choi Y, Shadel GS, Ghosh S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011; 472:476–480.

77. Geng J, Sun X, Wang P, Zhang S, Wang X, Wu H, Hong L, Xie C, Li X, Zhao H, Liu Q, Jiang M, Chen Q, Zhang J, Li Y, Song S, Wang HR, Zhou R, Johnson RL, Chien KY, Lin SC, Han J, Avruch J, Chen L, Zhou D. Kinases Mst1 and Mst2 positively regulate phagocytic induction of reactive oxygen species and bactericidal activity. Nat Immunol. 2015; 16:1142–1152.

78. Wi SM, Moon G, Kim J, Kim ST, Shim JH, Chun E, Lee KY. TAK1-ECSIT-TRAF6 complex plays a key role in the TLR4 signal to activate NF-kappaB. J Biol Chem. 2014; 289:35205–35214.

79. Lei CQ, Zhang Y, Li M, Jiang LQ, Zhong B, Kim YH, Shu HB. ECSIT bridges RIG-I-like receptors to VISA in signaling events of innate antiviral responses. J Innate Immun. 2015; 7:153–164.

80. Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011; 469:221–225.

81. Khan M, Syed GH, Kim SJ, Siddiqui A. Hepatitis B Virus-Induced Parkin-Dependent Recruitment of Linear Ubiquitin Assembly Complex (LUBAC) to Mitochondria and Attenuation of Innate Immunity. PLoS Pathog. 2016; 12:e1005693.

82. Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013; 501:512–516.

83. Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, Ramanujan VK, Wolf AJ, Vergnes L, Ojcius DM, Rentsendorj A, Vargas M, Guerrero C, Wang Y, Fitzgerald KA, Underhill DM, Town T, Arditi M. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012; 36:401–414.

84. Bruey JM, Bruey-Sedano N, Luciano F, Zhai D, Balpai R, Xu C, Kress CL, Bailly-Maitre B, Li X, Osterman A, Matsuzawa S, Terskikh AV, Faustin B, Reed JC. Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell. 2007; 129:45–56.

85. Bulua AC, Simon A, Maddipati R, Pelletier M, Park H, Kim KY, Sack MN, Kastner DL, Siegel RM. Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS). J Exp Med. 2011; 208:519–533.

86. Kim HJ, Kim CH, Ryu JH, Kim MJ, Park CY, Lee JM, Holtzman MJ, Yoon JH. Reactive oxygen species induce antiviral innate immune response through IFN-lambda regulation in human nasal epithelial cells. Am J Respir Cell Mol Biol. 2013; 49:855–865.

87. Yang CS, Yuk JM, Kim JJ, Hwang JH, Lee CH, Kim JM, Oh GT, Choi HS, Jo EK. Small heterodimer partner-targeting therapy inhibits systemic inflammatory responses through mitochondrial uncoupling protein 2. PLoS One. 2013; 8:e63435.

88. Park J, Min JS, Kim B, Chae UB, Yun JW, Choi MS, Kong IK, Chang KT, Lee DS. Mitochondrial ROS govern the LPS-induced pro-inflammatory response in microglia cells by regulating MAPK and NF-kappaB pathways. Neurosci Lett. 2015; 584:191–196.

89. Mills EL, Kelly B, Logan A, Costa AS, Varma M, Bryant CE, Tourlomousis P, Dabritz JH, Gottlieb E, Latorre I, Corr SC, McManus G, Ryan D, Jacobs HT, Szibor M, Xavier RJ, Braun T, Frezza C, Murphy MP, O'Neill LA. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell. 2016; 167:457–470.

90. Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, Oyabu J, Murakawa T, Nakayama H, Nishida K, Akira S, Yamamoto A, Komuro I, Otsu K. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012; 485:251–255.

91. Sun S, Sursal T, Adibnia Y, Zhao C, Zheng Y, Li H, Otterbein LE, Hauser CJ, Itagaki K. Mitochondrial DAMPs increase endothelial permeability through neutrophil dependent and independent pathways. PLoS One. 2013; 8:e59989.

92. West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, Kaech SM, Smiley JR, Means RE, Iwasaki A, Shadel GS. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015; 520:553–557.

93. Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011; 12:222–230.

94. Won JH, Park S, Hong S, Son S, Yu JW. Rotenone-induced impairment of mitochondrial electron transport chain confers a selective priming signal for NLRP3 Inflammasome Activation. J Biol Chem. 2015; 290:27425–27437.

95. Misawa T, Takahama M, Kozaki T, Lee H, Zou J, Saitoh T, Akira S. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat Immunol. 2013; 14:454–460.

96. Gottlieb RA, Carreira RS. Autophagy in health and disease. 5. Mitophagy as a way of life. Am J Physiol Cell Physiol. 2010; 299:C203–C210.

97. Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012; 13:255–263.

98. Okamoto K, Kondo-Okamoto N. Mitochondria and autophagy: critical interplay between the two homeostats. Biochim Biophys Acta. 2012; 1820:595–600.

99. Rodgers MA, Bowman JW, Liang Q, Jung JU. Regulation where autophagy intersects the inflammasome. Antioxid Redox Signal. 2014; 20:495–506.

100. Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A. 2009; 106:2770–2775.

101. O'Sullivan TE, Johnson LR, Kang HH, Sun JC. BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity. 2015; 43:331–342.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download