Abbreviations
BM
cDCs
CDP
CHO cells
DCs
EGFP
Flt3L
IRES
Lin
MCS
MDP
miRNA
pDCs
pri-miRNA
UTR
Journal List > Immune Netw > v.16(1) > 1033535
BM
cDCs
CDP
CHO cells
DCs
EGFP
Flt3L
IRES
Lin
MCS
MDP
miRNA
pDCs
pri-miRNA
UTR
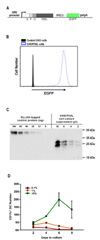 | Figure 1Culture of BM cells with CHO/Flt3L-conditioned medium produces DCs in vitro. (A) Diagram for the expression construct that encodes soluble FLAG and OLLAS tagged mouse Flt3L gene with IRES and EGFP (SFO.Flt3L-IRES-EGFP). (B) CHO cells stably transfected with SFO.Flt3L-IRES-EGFP (CHO/Flt3L cells) were selected and cloned for the high expression of EGFP. (C) Concentration of mouse Flt3L protein in the supernatant from CHO/Flt3L cell culture was titrated using anti-OLLAS monoclonal antibody. (D) Time-course quantification of CD11c+ DCs per well for each culture condition containing 0.1~10% of Flt3L conditioned medium. |
 | Figure 2Preliminary expression profiles of candidate miRNAs in Flt3L-cultured DC subsets. (A) Gating strategies for pDCs and cDCs present in BM culture with Flt3L for 8 days. (B) Normalized expression levels of candidate miRNAs in DC subsets isolated from BM culture with Flt3L. (C) Relative expression of individual candidate miRNAs between pDCs and cDCs isolated from BM culture with Flt3L. |
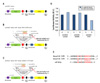 | Figure 3Regulation of gene expression by miR-124 via direct binding to 3'UTR of target transcript. Diagrams of the luciferase reporter vector pmirGLO constructs encoding (A) no insert, i.e., control, or 3'UTR from (B) TCF4 and (C) Zbtb46. (D) Histogram of normalized luciferase activities obtained from HeLa cells co-transfected with the respective reporter constructs and miR-124 mimic or negative control. Representative results are shown from 3 independent experiments. (E) Predicted binding site of miR-124 in the 3'UTRs of mouse and human TCF4. |
 | Figure 4High expression of miR-124 in cDC1 cells from BM culture with Flt3L. (A) Gating and sorting strategies for pDC, cDC1, and cDC2 cells from BM culture with Flt3L. (B) Relative expression of 3 transcription factors critical to DC development is determined amongst different DC subsets by real-time RT-PCR. Representative results are shown from 2 independent experiments. (C) Relative expression of miR-124 is assessed amongst different DC subsets by real-time RT-PCR. Data from 3 independent experiments are presented in histogram. Error bars indicate mean±SEM across all samples from 3 independent experiments. *p≤0.05; **p≤0.01; ***p≤0.001. |
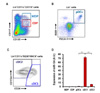 | Figure 5Prominent expression of miR-124 in cDC1 cells in BM. Gating strategies for (A) MDP, CDP, (B) pDC, (C) cDC1, and cDC2 cells in BM. (D) Relative expression of miR-124 is assessed amongst different progenitors and DC subsets by real-time RT-PCR. Representative results are shown from 3 independent experiments. ***p≤0.001. |
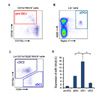 | Figure 6Elevated expression of miR-124 in cDC1 cells in spleen. Gating strategies for (A) pre-DC, (B) pDC, (C) cDC1, and cDC2 cells in spleen. (D) Relative expression of miR-124 is evaluated amongst different DC precursor and subsets by real-time RT-PCR. Representative results are shown from 3 independent experiments. ***p≤0.001. |
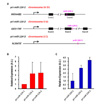 | Figure 7All three primary miR-124 genes are actively transcribed in DCs. (A) Genomic map of three pri-miR-124 genes on three different chromosomal locations are illustrated with their sequence information of GenBank accession numbers. (B) Relative expression of three pri-miR-124 genes is determined from cDC1 cells in BM by real-time RT-PCR. Results combined from 2 independent experiments are shown. (C) Relative expression of three pri-miR-124 genes is assessed from cDC1 cells in spleen by real-time RT-PCR. |
BM
cDCs
CDP
CHO cells
DCs
EGFP
Flt3L
IRES
Lin
MCS
MDP
miRNA
pDCs
pri-miRNA
UTR