Abstract
Exogenous nucleic acids induce an innate immune response in mammalian host cells through activation of the retinoic acid-inducible gene I (RIG-I). We evaluated RIG-I protein for RNA binding and ATPase stimulation with RNA ligands to investigate the correlation with the extent of immune response through RIG-I activation in cells. RIG-I protein favored blunt-ended, double-stranded RNA (dsRNA) ligands over sticky-ended dsRNA. Moreover, the presence of the 5'-triphosphate (5'-ppp) moiety in dsRNA further enhanced binding affinity to RIG-I. Two structural motifs in RNA, blunt ends in dsRNA and 5'-ppp, stimulated the ATP hydrolysis activity of RIG-I. These structural motifs also strongly induced IFN expression as an innate immune response in cells. Therefore, we suggest that IFN induction through RIG-I activation is mainly determined by structural motifs in dsRNA that increase its affinity for RIG-I protein and stimulate ATPase activity in RIG-I.
The appearance of viral nucleic acids in the mammalian cytoplasm triggers the antiviral defense mechanism by activating innate nucleic acid receptors such as retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), laboratory of physiology and genetics 2 (LGP2), and toll-like receptors (TLRs) (123). These cytosolic nucleic acid receptors serve as pattern recognition receptors (PRRs) of the innate immune system against pathogen-associated molecular patterns (PAMPs). Among these nucleic acid receptors, RIG-I is critical for sensing viral RNA and DNA in the cytoplasm of mammalian host cells. RIG-I belongs to the DExD/H family of helicases, which are characterized by both nucleic acid-binding and ATP hydrolysis activities (456). Activation of RIG-I by the binding of exogenous RNA present in the cytoplasm invokes antiviral responses including type I interferon (IFN) expression, inflammasome activation, and proapoptotic signaling (17).
RIG-I contains two N-terminal tandem caspase activation and recruitment domains (CARD), a central helicase superfamily II (SF2) ATPase domain, and a C-terminal regulatory domain (RD). Several reports have shown that C-terminal RDs recognize distinct RNA patterns such as single-stranded (ss) or double-stranded RNA (dsRNA) ends containing either 5'-triphosphate (5'-ppp) or 5'-diphosphate (5'-pp) moieties (789). These patterns are viral RNA signature motifs and are believed to be related to the extent of the type I IFN immune response in cells (8). Following ligand RNA recognition, RIG-I initiates an intracellular signaling cascade leading to the production of IFNs and inflammatory cytokines (9). If RIG-I undergoes conformational change arising from the binding of ligand RNAs, two CARD modules of RIG-I are sterically displaced from its inhibited form (610) and become poly-ubiquitinated (11). The activated CARDs of RIG-I cause polymerization of mitochondrial antiviral-signaling protein (MAVS) (12) and trigger activation of interferon regulatory factor 3 (IRF3). IRF3 induces interferon α/β (IFN-a/b) and interferon-stimulated genes (ISGs), which are key to innate immune defense (13).
Recently, our group demonstrated that RIG-I favors 5'-ppp RNA ligands over RNA ligands that lack the 5'-ppp moiety (i.e., HO-RNA), and the 5'-ppp moiety of RNA strands invokes elevated induction of type I IFN in cells that express RIG-I (7). However, apart from the RNA-binding activities of RIG-I the exact roles of ATP-binding and hydrolysis in activating RIG-I for immune signaling have not been clearly defined. Without an SF2 ATPase domain in RIG-I, overexpression of the two CARD modules alone is sufficient for innate immune signaling (14). The ATPase domain is an ATP-dependent dsRNA translocase that allows multiple loading of RIG-I on dsRNA (15). Recently, it has been suggested that ATP hydrolysis by RIG-I prevents recognition of endogenous RNA without invoking unintentional signaling through prolonged RNA binding (16).
In this study, we investigated the roles of ATP hydrolysis and RNA binding by RIG-I in the activation of innate immunity through the induction of type 1 interferon in cells. We found that RNA ligands that were blunt-ended and had terminal 5'-ppp moieties bound strongly to RIG-I and stimulated ATP hydrolysis activity in RIG-I. Type I interferon induction in lung epithelial cells was mostly activated by dsRNA with blunt ends and 5'-ppp termini. Our results suggest that the presence of the triphosphate moiety at the blunt-ended 5'-termini in dsRNA is a major determinant for type I interferon response as innate immunity.
Recombinant human full-length RIG-I (1–925) protein harboring a hexa-histidine tag at the N-terminal was expressed in Escherichia coli pET-22b expression vectors (Novagen, Darmstadt, Germany) that were introduced into BL21 (DE3) cells (Novagen). The cells were grown in a shaking incubator at 37℃ in Luria broth medium supplemented with 50 µg/mL ampicillin. Protein expression was induced by the addition of 0.5 mM isopropyl-d-1-thiogalactopyranoside (IPTG). The cells were then further incubated at 16℃ for 24 h. After harvesting, resuspended cells were disrupted by sonication. All recombinant proteins were purified with Ni fast protein liquid chromatography (FPLC) using standard protocols (GE Healthcare, Piscataway, NJ, USA) (17).
The ssRNA and single-stranded DNA (ssDNA) substrates were chemically synthesized (ST pharm Co. Ltd. Seoul, Korea). The 5'-ppp ssRNAs were synthesized by in vitro transcription using T7 polymerase, as described previously (18). For generating duplexes, RNA or DNA oligonucleotides were mixed in annealing buffer (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 1 mM EDTA), boiled at 85℃ for 3 min, and incubated at 25℃ for 20 min. Where indicated, oligonucleotides were 5'-phosphorylated using T4 polynucleotide kinase (TaKaRa, Tokyo, Japan). Yeast tRNA and polyinosinic:polycytidylic acid (poly I:C) nucleotides were purchased from Sigma (St. Louis, MO, USA).
The ATPase assay reaction mixtures contained buffer (20 mM Tris-HCl, pH 7.5, 8 mM DTT, 5 mM MgCl2, 10 mM KCl), 4% (w/v) glycerol, and the specified RNA (1 µM) plus a trace amount of [γ-32P]ATP (2 nM, GE Healthcare) mixed with nonradioactive ATP (200 µM) and 200 nM RIG-I. The reactions were initiated by the addition of enzyme and the reactants were incubated at 37℃. At 3 min intervals, the reaction was stopped by the addition of 1 µL of 4 M formic acid to each aliquot (1.5 µL) of reaction mixture. The quenched reaction aliquot (1 µL) was blotted on polyethyleneimine-cellulose for thin-layer chromatography (Macherey-Nagel, Düren, Germany) and developed in 0.4 M potassium phosphate (pH 3.4). Unreacted ATP and product Pi were separated and quantified using a Cyclone Phosphorimager (Packard Instrument Co., Inc., Meriden, CT, USA).
Various amounts (0, 5, 10, 15, 20, 25, 30, 60, and 80 pmol) of recombinant RIG-I protein were mixed with 20 pmol of RNA oligonucleotides in 25 µL of binding buffer (20 mM Tris-HCl, pH 7.5, 1.5 mM MgCl2, 1.5 mM DTT) for 20 min at 37℃. The reaction mixture was then applied to a 2% agarose gel (containing Tris/borate/EDTA buffer) that was subsequently stained, after which the RNAs were visualized using a UV transilluminator. To detect the proteins, the gel was stained with Coomassie Brilliant Blue.
The carcinoma human alveolar basal epithelial cell line A549 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). The A549 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Hyclone Laboratories Inc., Logan, UT, USA) supplemented with 100 U of penicillin, 100 mg of streptomycin, and 10% fetal bovine serum (FBS; Gibco-BRL, Grand Island, NY, USA) at 37℃ in 5% CO2. Transfection of RNA oligonucleotides was conducted using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions.
A549 cells were seeded onto a 24-well plate (1.0×105) and incubated for 12 h, after which they were transfected with the reporter constructs, pGL-IFN-β (0.2 µg) and pRL-TK (0.1 µg), as used previously (19). The cells were further incubated for 8 h, after which they were transfected with the designated amount of each RNA oligonucleotide. Yeast tRNA and poly I:C RNA were used as negative and positive controls, respectively. After 16 h, the cells were collected and disrupted with Passive Lysis buffer (Promega, Madison, WI, USA) and the cell extracts were subjected to a luciferase assay using a dual-luciferase assay system (Promega). The levels of luminescence from firefly and Renilla luciferases were measured using a VICTOR X3 Multilabel Plate Reader (PerkinElmer, Waltham, MA, USA), and the ratio of firefly luciferase to Renilla luciferase was calculated.
To determine the binding affinity between RNA substrates and RIG-I protein, we carried out an electrophoretic mobility shift assay (EMSA) for monitoring RNA:protein complex formation. We examined several types of duplex oligonucleotides comprising dsRNA with either blunt or sticky ends and RNA:DNA hybrids (Table I). These partial duplex RNA or RNA:DNA hybrids contained 5'-hydroxyl moieties instead of triphosphate moieties at the 5'-terminal. Fixed quantities of the RNA ligands were mixed with varying amounts of RIG-I protein (0, 5, 10, 15, 20, 25, 30, 60, and 80 pmol). The results on 2% agarose gel showed that the 3'-overhang RNA substrate (47R/25RI) band was more rapidly shifted at low concentrations of RIG-I than the other types of RNA substrate (5'-overhang and double overhang) (Fig. 1A). This indicates that RIG-I proteins have a high affinity for duplex RNA molecules with blunt-ended 5'-termini. In addition, the RNA:DNA hybrid substrates (47R/25DI and 47R/25DII) had a lower binding affinity for RIG-I than the dsRNA substrates (47R/25RI, 47R/25RII, and 47R/25RIII). Therefore, a duplex RNA with 5'-blunt ends is the favored ligand for RIG-I binding; the RNA:DNA duplex did not support RIG-I binding, even in the presence of 5'-blunt-ended termini.
We next investigated the effect of the 5'-ppp moiety on the binding efficacy of RIG-I to the 5'-blunt-ended duplex of RNA or RNA:DNA. As shown in Fig. 1B, the presence of 5'-ppp in the duplex RNA or RNA:DNA significantly affected binding affinity to RIG-I; the amount of duplex RNA or RNA:DNA that were bound to RIG-I was apparently enhanced in the presence of 5'-ppp in the duplex nucleic acid ligands. These results suggest that RIG-I prefers duplex RNA with blunt-ended 5'-ppp termini for effective binding.
We hypothesized that the binding affinity of RIG-I for RNA substrates is correlated with ATP hydrolysis activity. Therefore, we next examined the RNA ligand structure preference of RIG-I for ATP hydrolysis stimulation. RIG-I protein was incubated at room temperature for various periods with ATP spiked with [γ-32P]ATP after the addition of several RNA substrates. The reaction mixture was subjected to polyethyleneimine-cellulose thin-layer chromatography, and the reaction products (ADP and Pi) were resolved. ATP hydrolyzed by RIG-I was quantified by measuring the radioactivity of the products and the signal intensity was plotted (Fig. 2A). The results revealed that the ATP hydrolysis activity of RIG-I was stimulated by dsRNA ligands. The dsRNA ligands (47R/25RI and 47/25RII) that contained a 25-bp duplex region and a 22-nt single-stranded tail stimulated ATPase activity equally, regardless of the position of the single-stranded tail. However, the presence of blunt ends in the dsRNA was important for the stimulation of ATPase activity; the dsRNA ligand (47R/25RIII) containing the 25-bp duplex region and 11-nt tails at both ends showed lower ATPase activity than the other dsRNAs.
To determine whether the presence of 5'-ppp in RNA affects the ATPase activity of RIG-I, time-dependent hydrolysis of ATP by RIG-I was monitored in the presence of ssRNAs with either 5'-ppp or 5'-OH moieties at the termini (Fig. 2B). ATP hydrolysis by RIG-I was significantly stimulated by ssRNA with 5'-ppp, whereas ssRNA with a hydroxyl group at the 5' terminus showed slower ATPase activity compared with 5'-ppp ssRNA. Neither ssDNA without 5'-ppp nor shorter ssRNA stimulated ATP hydrolysis activity to the degree observed in ssRNA with 5'-ppp. When the ATP hydrolysis kinetics were compared, ATP hydrolysis activity was stimulated to a greater extent by blunt-ended dsRNA than by 5'-ppp ssRNA. Taken together, these results suggest that RIG-I prefers double-stranded RNA with a blunt end and/or 5'-ppp at the terminus for ATP hydrolysis.
The results of RNA binding and ATP hydrolysis by RIG-I prompted us to hypothesize that interferon (IFN) production via RIG-I would be strongly enhanced with the exogenous RNA ligands preferred by RIG-I. To test the hypothesis, various types of dsRNA and RNA:DNA hybrids were transfected into lung epithelial cells (A549 cells), and IFN expression levels were quantified using the dual luciferase assay (Fig. 3). Previously, we observed that RNAs containing the 5'-ppp moiety significantly increased the expression of type I IFN compared with their RNA counterparts without the 5'-ppp moiety in A549 cells through a RIG-I-mediated innate immune response (7). In addition, it has been recently reported that the presence of 5'-triphosphate in duplex RNA facilitate conformational change of RIG-I to form dimers by exposing CARD domain through interaction with positively charged residues of RIG-I such as lysine or histidine, which can stimulate IFN-β induction pathway in cells. However, absence of 5'-ppp in duplex RNA ligands is unable to cause swiveling of CARD domain, resulting in the absence of IFN-β induction (20). Therefore, we also prepared duplex nucleic acid ligands containing RNA strands with 5'-ppp moieties. We first examined the induction of IFN production with dsRNAs and RNA:DNA hybrids, which are devoid of the 5'-ppp moiety. None of the ligands stimulated IFN promoter reporter gene activity, regardless of the ligand transfection dosage. In contrast, RNAs containing the 5'-ppp moiety significantly increased the expression of type I IFN compared with their RNA counterparts without the 5'-ppp moiety in the A549 cells. Thus, the presence of ppp at the 5' terminus in the nucleic acid ligand is a major determining factor for IFN induction of the innate immune response via the RIG-I pathway. Of the 5'-ppp RNA ligands that stimulated IFN expression, the 47R/25RI double-stranded RNA showed greater IFN expression than any other type of nucleic acid ligand. Taken together, these results suggest that RIG-I favors a blunt-ended dsRNA with a 5'-ppp moiety at the terminus as a nucleic acid ligand for innate immune response.
In conclusion, the presence of 5'-ppp and a 5' blunt end in the dsRNA substrate enhances the binding affinity of RIG-I to the dsRNA ligand as well as the ATPase activity of RIG-I. These structural motifs provide a basis for IFN expression as an innate immune response via the RIG-I pathway. Furthermore, the presence of 5'-ppp in the dsRNA strongly stimulates IFN induction via RIG-I in lung epithelial cells, in which intracellular RIG-I is abundant. Thus, the present study suggests that IFN induction as an innate immune response via the RIG-I pathway is positively correlated with structural motifs in dsRNA that dictate both high affinity for RIG-I and stimulation of ATPase activity by RIG-I.
Figures and Tables
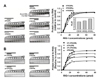 | Figure 1Binding affinities of various types of duplex nucleic acid ligands to RIG-I protein. (A) Increasing amounts of RIG-I protein were incubated with each duplex nucleic acid ligand; the dsRNA ligands included 3'-end overhang (47R/25RI), 5'-end overhang (47R/25RII), double overhang (47R/25RIII)' and RNA:DNA hybrids including 3'-end overhang (47R/25DI) and 5'-end overhang (47R/25DII). Nucleic acid ligands complexed with the RIG-I protein appeared as slowly migrating bands on 2% agarose gel after the electrophoretic mobility shift assay (EMSA). The nucleic acid band intensities were quantitated by ImageJ software, and the extent of nucleic acid ligands bound to the RIG-I protein were plotted as normalized values relative to the control (No RIG-I protein). Each curve was fitted to the Hill equation; Y=A×Lh/(Bh+Lh), where Y represents for relative fraction of RNA ligands bound to RIG-I at each RNA ligand concentration (L), A represents for the amount of RNA ligands complexed to RIG-I at equilibrium, and B and h represents apparent dissociation constant (i.e. Kd) and the Hill coefficient, respectively. Each apparent Kd (in µM) value was obtained from the fitting of the curve to the above equation using Sigma Plot, and represented as the bar graph (inset). (B) Increasing amounts of RIG-I protein were incubated with double-stranded nucleic acids with 3'-end overhang and 5'-end triphosphate (3P-47R/25RI and 3P-47R/25DI) or 5'-end-OH (47R/25RI and 47R/25DI). After the EMSA, the migration of the nucleic acid ligands was visualized on the gel. The upper band represents nucleic acids bound to RIG-I, whereas the bottom band shows unbound free nucleic acid ligands. The nucleic acid band intensities were quantitated by ImageJ software, and the extent of nucleic acid ligands bound to the RIG-I protein were plotted as normalized values relative to the control (No RIG-I protein). Each curve was fitted to the Hill equation; the amplitudes of each curve were different at equilibrium. |
 | Figure 2Stimulation of ATP hydrolysis by RIG-I with various types of nucleic acid ligands. ATP (200 M) was mixed with RIG-I (20 nM) in the presence of each nucleic acid ligand (0.5 nM). (A) ATP hydrolysis by RIG-I was monitored over time in the presence of dsRNAs including 3'-end overhang (47R/25RI), 5'-end overhang (47R/25RII), and double overhang (47R/25RIII). The thin-layer chromatography plates show the progress of the ATP hydrolysis reaction, in which ATP hydrolysis products (radioactive inorganic phosphate, Pi*) and radioactive substrates ATP* were resolved and quantified. (B) The ATPase activity of RIG-I over time was monitored in the presence of ssRNAs including 5'-ppp RNA (3P-47R) and ssRNA or ssDNA with a 5'-end hydroxyl group (47R, 25RI, and 25DI). |
 | Figure 3Interferon induction in lung epithelial cells with various types of nucleic acid ligands through RIG-I activation. A549 cells were transfected with the 5'-ppp or 5'-HO RNA substrates or the RNA:DNA hybrid substrate. A549 cells containing the pGL3-IFNβ reporter gene were transfected with each duplex RNA ligand at two different concentrations (10 nM or 50 nM). The luciferase reporter assay was conducted at 18 h post-transfection with each RNA ligand. Activation of the IFN-β promoter with the exogenous RNA ligand is shown in the graph as the fold-index relative to a mock transfection control (transfection reagent only). Transfections with yeast tRNA and poly I:C were performed for negative and positive controls, respectively. The unit fold was set to the IFN expression observed in the mock transfection. The values shown are the means±S.D. of triplicate experiments. |
Table I
Sequences of single-stranded oligonucleotides used in this study (Top panel). Schematic diagrams of duplex nucleic acids such as dsRNAs and RNA:DNA hybrids, which are used in this study, and composing oligonucleotides. Black colored bar and grey colored bar represents RNA and DNA, respectively (Bottom panel).

| Name | Duplex RNA or RNA : DNA | Annealing oligonucleotides |
|---|---|---|
| 47R/25RI |
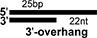
|
1+2 |
| 47R/25RII |
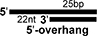
|
1+3 |
| 47R/25RIII |

|
1+4 |
| 47R/25DI |

|
1+5 |
| 47R/25DII |
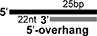
|
1+6 |
ACKNOWLEDGEMENTS
This work was supported by a grant from Ministry of Health and Welfare, Republic of Korea (HI15C2917).
References
1. Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol. 2015; 33:257–290.

2. Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol. 2014; 32:461–488.

3. Thompson AJ, Locarnini SA. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol Cell Biol. 2007; 85:435–445.

4. Gee P, Chua PK, Gevorkyan J, Klumpp K, Najera I, Swinney DC, Deval J. Essential role of the N-terminal domain in the regulation of RIG-I ATPase activity. J Biol Chem. 2008; 283:9488–9496.

5. O'Neill LA, Bowie AG. The powerstroke and camshaft of the RIG-I antiviral RNA detection machine. Cell. 2011; 147:259–261.
6. Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M Jr, Patel SS, Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011; 479:423–427.

7. Baek SE, Kim H, Kim KB, Yoon S, Choe J, Suh W, Jeong YJ, Cho YH, Kim DE. Dual effects of duplex RNA harboring 5'-terminal triphosphate on gene silencing and RIG-I mediated innate immune response. Biochem Biophys Res Commun. 2015; 456:591–597.

8. Ranjith-Kumar CT, Murali A, Dong W, Srisathiyanarayanan D, Vaughan R, Ortiz-Alacantara J, Bhardwaj K, Li X, Li P, Kao CC. Agonist and antagonist recognition by RIG-I, a cytoplasmic innate immunity receptor. J Biol Chem. 2009; 284:1155–1165.

9. Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T, Hartmann G, Patel DJ. Structural and functional insights into 5'-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010; 17:781–787.

10. Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011; 147:423–435.

11. Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007; 446:916–920.

12. Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, Walz T, Penczek PA, Hur S. Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol Cell. 2014; 55:511–523.

13. Foy E, Li K, Wang C, Sumpter R Jr, Ikeda M, Lemon SM, Gale M Jr. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003; 300:1145–1148.

14. Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004; 5:730–737.

15. Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5'-triphosphate-dependent translocase on double-stranded RNA. Science. 2009; 323:1070–1074.

16. Lassig C, Matheisl S, Sparrer KM, de Oliveira Mann CC, Moldt M, Patel JR, Goldeck M, Hartmann G, Garcia-Sastre A, Hornung V, Conzelmann KK, Beckmann R, Hopfner KP. ATP hydrolysis by the viral RNA sensor RIG-I prevents unintentional recognition of self-RNA. Elife. 2015; 4:e10859.
17. Lee SY, Jung HY, Kim TO, Im DW, You KY, Back JM, Kim Y, Kim HJ, Shin W, Heo YS. Cloning, purification, crystallization and preliminary X-ray crystallographic analysis of the N-terminal domain of DEAD-box RNA helicase from Staphylococcus aureus strain Mu50. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010; 66:1674–1676.

18. Lee B, Kim KB, Oh S, Choi JS, Park JS, Min DH, Kim DE. Suppression of hepatitis C virus genome replication in cells with RNA-cleaving DNA enzymes and short-hairpin RNA. Oligonucleotides. 2010; 20:285–296.

19. Zhu FX, King SM, Smith EJ, Levy DE, Yuan Y. A Kaposi's sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc Natl Acad Sci U S A. 2002; 99:5573–5578.

20. Lee MK, Kim HE, Park EB, Lee J, Kim KH, Lim K, Yum S, Lee YH, Kang SJ, Lee JH, Choi BS. Structural features of influenza A virus panhandle RNA enabling the activation of RIG-I independently of 5'-triphosphate. Nucleic Acids Res. 2016; DOI: 10.1093/nar/gkw525.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download