Abstract
CCR3 is a chemokine receptor that mediates the accumulation of allergic inflammatory cells, including eosinophils and Th2 cells, at inflamed sites. The regulatory sequence of the CCR3 gene, contains two Runt-related transcription factor (RUNX) 1 sites and two PU.1 sites, in addition to a functional GATA site for transactivation of the CCR3 gene. In the present study, we examined the effects of the cis-acting elements of RUNX1 and PU.1 on transcription of the gene in EoL-1 eosinophilic cells and Jurkat T cells, both of which expressed functional surface CCR3 and these two transcription factors. Introduction of RUNX1 siRNA or PU.1 siRNA resulted in a modest decrease in CCR3 reporter activity in both cell types, compared with transfection of GATA-1 siRNA. Cotransfection of the two siRNAs led to inhibition in an additive manner. EMSA analysis showed that RUNX1, in particular, bound to its binding motifs. Mutagenesis analysis revealed that all point mutants lacking RUNX1- and PU.1-binding sites exhibited reduced reporter activities. These results suggest that RUNX1 and PU.1 participate in transcriptional regulation of the CCR3 gene.
The CC chemokine receptor CCR3 plays a major role in the development of allergic diseases by mediating the recruitment of allergic cells to inflamed sites (1). Eosinophils are primarily found in the inflammatory infiltrate, but Th2 cells also have critical roles in the allergic pathology (2). These cells express CCR3 on their cell surfaces (3). CCR3 appears to be regulated in a temporal manner during phenotypic differentiation of eosinophils and is expressed along with IL-5Rα at a basal level in eosinophil progenitors (4). After commitment of eosinophils, CCR3 expression gradually increases during differentiation and reaches its highest level at the terminal stage both in vivo (5) and in vitro (6).
CCR3 expression is mainly regulated at the transcription level. A region spanning exon 1 and proximal intron 1 of this gene have been shown to retain the critical sequence for transcriptional control (789). This sequence harbors multiple GATA sites for the zinc finger transcription factor GATA-1. We have previously mapped the functional GATA site within exon 1 of this gene, to which GATA-1 binds with high affinity. Introduction of GATA-1 siRNA or dominant negative GATA-1 resulted in a significant reduction in CCR3 reporter activity (6). Although GATA-1 acts as a key regulator of CCR3 gene transcription, it alone is unlikely to be sufficient for full transactivation of the reporter, since a point mutant lacking the functional GATA site exhibits 50~60% reduced transcription. Close examination reveals that, in addition to GATA sites, the regulatory sequence of the CCR3 gene contains two cis-acting elements of the transcription factors Runt-related transcription factor 1 (RUNX1) and PU.1. PU.1 is selectively expressed in B lymphocytes, granulocytes, and monocytes and is required for eosinophil development, regulating an eosinophil-specific gene in corporation with GATA-1 and CCAAT/enhancer binding protein alpha (1011121314). It contains various distinct functional domains, namely an Ets domain that recognizes the DNA sequence harboring the core GGAA motif (15). RUNX1 is required for generation of hematopoietic lineages including myeloid lines (1617). Genetically modified mice that do not express RUNX1 demonstrate a dramatic decrease in basophils but normal numbers of neutrophils and eosinophils, suggesting that RUNX1 plays a role in differentiation of basophils (18). RUNX1 binds to DNA in a sequence-specific manner, recognizing a TGTGGT consensus binding site (19). Given the intimate relationship of RUNX1 and PU.1 to hematopoietic components and the presence of their cis-acting factors in the key regulatory region of the CCR3 gene, we investigated their involvement in CCR3 transcription.
Jurkat cells were purchased from the Korean Cell Line Bank (Seoul, Korea). EoL-1 cells were kindly provided by Yun-Jae Jung (Gacheon University, Incheon, Korea). Jurkat and EoL-1 cells were maintained in RPMI medium (Welgene, Seoul, Korea) supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (100 µg/ml).
Total mRNAs were extracted from cell lines using TRI reagent (Molecular Research Center, Cincinnati, OH, USA). First-strand cDNA was synthesized from 4 µg total RNA using SuperScript II Reverse Transcriptase (Invitrogen Life Technologies) in a 20 µl reaction containing random primers, deoxynucleotide triphosphates (0.5 mM), MgCl2 (2.5 mM), and DTT (5 mM). Reverse transcription was performed at 42℃ for 1 h, followed by heat inactivation at 70℃ for 15 min. The synthesized cDNA was amplified for 30 cycles with Ex DNA polymerase (TAKARA, Shiga, Japan). The following primers were used in the amplification: CCR3 forward 5'-ATGCTGGTGACAGAGGTGAT-3' and reverse 5'-AGGTGAGTGTGGAAGGCTTA-3'; GAPDH forward 5'-CGTCTTCACCACCATGGAGA-3' and reverse 5'-CGGCCATCACGCCACAGTTT-3.'
Cells were lysed in RIPA buffer (50 mM Tris-Cl [pH 7.4], 0.1% NaN3, 1% Nonidet P-40, 0.25% sodium deoxycholate, 1 mM EDTA, 1 mM Na3VO4, 1 mM NaF, and protease inhibitor mixture) supplemented with 0.4 M NaCl. Lysates were centrifuged, and the resulting supernatants were subjected to Western blot analysis. Thirty micrograms of the cell lysates were resolved with SDS-PAGE and then transferred to polyvinylidene difluoride membranes. After the membranes were blocked with 5% nonfat dry milk, they were probed with anti-RUNX1 (N-20, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-PU.1 (T-21, Santa Cruz Biotechnology), and anti-GAPDH Ab (Santa Cruz Biotechnology). The membranes were incubated with an anti-goat HRP-conjugated Ab (Santa Cruz Biotechnology) for RUNX1 and an anti-rabbit HRP-conjugated Ab (Cell Signaling Technology, Beverly, MA, USA) for PU.1. Immunostained proteins were visualized using an ECL detection system (Amersham Biosciences, Buckinghamshire, UK).
Cells were fixed in 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO, USA) for 15 min at 4℃, washed with 2% BSA in PBS, and then stained with PE-conjugated anti-human CCR3 Ab (R&D Systems, Minneapolis, MN, USA) or isotype matched Ab (BD Pharmingen, San Diego, CA, USA). Stained cells were analyzed using a flow cytometer, and data analysis was performed using CellQuest Software (Becton Dickinson, San Jose, CA, USA).
Chemotaxis assays were performed in 48-well Boyden chambers (Neuro Probe, Gaithersburg, MD, USA). EoL-1 and Jurkat cells were washed and resuspended in chemotaxis medium containing 1% BSA and 2 mM HEPES. Assays were performed in triplicates using 2×105 cells/well with a 5-µm-pore size PVPF membrane (Osmonics, Minnetonka, MN, USA). A 27 µl aliquot of indicated concentrations of eotaxin was added to the lower chamber. A filter was overlaid and then 50 µl of the cell suspension was added to the upper chambers. The chamber was incubated at 37℃ in 5% CO2 for 3 h. Then, cells which migrated to the lower chamber were counted. Chemotaxis was expressed as a chemotaxis index.
Small interfering RNA (siRNA)-dependent knockdown experiments were conducted using ON-TARGETplus SMARTpool siRNA for RUNX1, PU.1, GATA-1, and non-targeting (scrambled) control pools (Dharmacon, Chicago, IL, USA). Briefly, EoL-1 and Jurkat cells were cotransfected with siRNA and the CCR3 reporter using Amaxa 4D-nucleofector (Lonza, Koln, Germany) or Lipofectamine 2000 Reagent (Invitrogen Life Technologies) and examined for luciferase activities.
A pGL3-based luciferase reporter construct (Promega, Madison, WI, USA) that harbored exon 1 (249 bp) and the proximal intron 1 (157 bp) of the CCR3 gene was previously described (6). RUNX1 and PU.1 sites were mutated using conventional overlap extension PCR with the primers listed in Table I. The RUNX1 site 5'-AGTGGT-3' was replaced with 5'-CTGTTG-3.' The PU.1 sites, which occurred in tandem repeat as 5'-GAGGAGAGGA-3,' were mutated to 5'-TCGGCTCGGC-3' and treated as a single site. Amplification was performed using Pyrobest Taq™ (TAKARA) in a 50 µl reaction. The resulting PCR products were cloned into the pGL3 vector, and their sequences were confirmed through DNA sequencing analysis. Cell lines were transfected with the reporter plasmids. Luciferase activity was measured using the Dual-Luciferase Reporter System (Promega), and transfection efficiency was normalized to Renilla luciferase activity from cotransfection with pRL-TK vector (Promega).
After lysis with RIPA buffer containing 0.15 M NaCl, the lysate was centrifuged. The pellet was resuspended in RIPA buffer containing 0.4 M NaCl. The supernatant was then used for EMSA analysis. Four micrograms of the nuclear extracts were incubated with 32P-labeled probe for 20 min in binding buffer (50 mM Tris-Cl [pH 7.5], 20% glycerol, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM DTT, 250 mM NaCl, and 0.25 mg/ml poly[dI-dC]·poly[dI-dC]). For supershift analysis, extracts were incubated with anti-RUNX1 (N-20, Santa Cruz Biotechnology), anti-PU.1 (T-21, Santa Cruz Biotechnology), or control antibodies (Sigma-Aldrich), followed by a radiolabeled probe. Reaction mixtures were analyzed through separation on 6% polyacrylamide gels. RUNX1 and PU.1 site probes are as follows: RUNX1 upstream site, 5'-TTTCAGGAGTGGTGACGCCT-3'; RUNX1 downstream site, 5'-AGCAGGTACCACTGGTCTTC; PU.1 site, 5'-GGAGGGAGGAGAGGAAGGTA-3.'
All statistical analyses were performed using the Microsoft Excel data analysis program or SPSS 12.0 software (SPSS, Inc., Chicago, IL, USA). An independent t test was used to compare relative reporter activities between the two groups. Comparisons of the activities in three or more independent groups were performed using one-way ANOVA, followed by post-hoc analysis. Differences with a p-value less than 0.05 were considered statistically significant. The results are expressed as mean±SEM.
We chose the two cell lines EoL-1 and Jurkat, which represent eosinophils and T cells, respectively. RT-PCR and FACS analyses showed that the two cell types constitutively expressed not only CCR3 mRNA but also surface CCR3 protein at their surfaces (Fig. 1A). The surface CCR3 protein was functional in both cell types, as evidenced by typical chemotactic responses to eotaxin, a CCR3 ligand (Fig. 1A). These two cells also expressed both RUNX1 and PU.1 proteins, as analyzed using RT-PCR (data not shown) and immunoblots (Fig. 1B).
We previously constructed a CCR3 reporter plasmid that spans exon 1 and the proximal intron 1 sequence (6), which includes two RUNX1 and two PU.1 sites. As EoL-1 and Jurkat cells expressed both RUNX1 and PU.1 transcription factors (Fig. 1B), we examined the involvement of RUNX1 and PU.1 in reporter activation. To this end, RUNX1 and PU.1 siRNAs were transfected into EoL-1 and Jurkat cells. GATA siRNA and scramble siRNA were used as positive and negative controls, respectively. GATA-1 siRNA reduced the reporter activity by 60% in both cell types, as observed in our previous report, while RUNX1 and PU.1 siRNAs reduced reporter activity by 22% and 28%, respectively (Fig. 2). An additive effect (40%) was observed upon cotransfection of RUNX1 and PU.1 siRNAs. This result indicates that RUNX1 and PU.1 participate in activation of CCR3 transcription, although they have a weaker effect than GATA-1.
The nucleotide sequence of the regulatory region of the CCR3 gene is shown in Fig. 3A. This region contains two RUNX1 sites in exon 1 and two tandem PU.1 sites in the proximal sequence of intron 1, in addition to the GATA site in exon 1 that has been demonstrated to be important for reporter activity (6). A point mutation in the functional GATA site (Construct 2 in Fig. 3B) resulted in a significant reduction in CCR3 reporter activity in EoL-1 and Jurkat cells, which is consistent with a previous report (20). Mutation in either of the two RUNX1 sites (constructs 3 and 4) resulted in a decrease in CCR3 reporter activity, with a smaller effect at the upstream RUNX1 site than the downstream RNUX1 site. Double mutations of one of the two RUNX1 sites along with the GATA site produced opposing results: one with mutation of the distal RUNX1 site (Construct 5) had reduced reporter activity with an additive effect of the GATA mutation, whereas the other with mutation of the proximal RUNX1 site (Construct 6) had increased activity. These RUNX1 sites constitute position-dependent positive and negative regulatory elements in the context of the major transcriptional regulatory element, the GATA site, thus contributing to fine-control of transcription of the CCR3 gene. Both functionally positive and negative interactions between RUNX1 and GATA factors have been exemplified in a previous report, in which aIIb integrin promoter in megakaryocytes is activated or repressed by a subtle difference in interaction of RUNX1 and GATA-1 (21).
The point mutant of the PU.1 sites (construct 7 in Fig. 3B) exhibited slightly but significantly reduced the reporter activity. The effect of PU.1 mutation (construct 7) was comparable to or slightly less than the effects of the RUNX1 mutations (constructs 3 and 4). A double mutation (construct 8) in the PU.1 site along with the GATA site yielded additively reduced activity. PU.1 is critical for normal granulocytic differentiation. Disruption of the PU.1 gene affects multiple hematopoietic lineages, including defects in granulocyte terminal differentiation, resulting in the loss of functionally mature neutrophils and eosinophils (111222). Furthermore, PU.1 activates transcription of the MBP gene, a representative eosinophil-specific gene. The MBP reporter is transactivated at a modest level by PU.1 alone but is synergistically transactivated by PU.1 and GATA-1. Removal of the PU.1 sites in the MBP promoter results in a modest decrease in reporter activity, while GATA mutations greatly reduced reporter activity (23). Our results are largely consistent with PU.1 and GATA-1 regulation of the MBP promoter. PU.1 siRNA and elimination of PU.1 sites modestly reduced the CCR3 reporter. A combined effect of PU.1 and GATA-1 was observed, although the effect was not synergistic, as evidenced by additive decreases upon elimination of both PU.1 and GATA sites (construct 8 in Fig. 3B) and the inclusion of PU.1 and GATA-1 siRNAs. PU.1 is definitely involved in CCR3 transactivation.
We next examined in vitro binding of RUNX1 and PU.1 to their putative elements. When nuclear extract from EoL-1 and Jurkat cells was reacted with each probe of the two RUNX1 sites, two DNA-protein complexes were formed. Incubation with anti-RUNX1 antibody induced formation of a supershifted band only with the fast-migrating species. The supershifted band overlapped with the slow migrating band that remained unchanged even in the presence of the anti-RUNX1 antibody. In contrast, the control antibody had no effect (Fig. 4). The DNA-protein complex was formed to a greater extent with the distal RUNX1 probe than that with the proximal RUNX1 probe. This result was opposite to that of reporter assays, in which the effect of the distal RUNX1 mutation was smaller than that of the proximal RUNX1 mutation. This discrepancy might be due to the fact that the EMSA probes used only include the RUNX1 sites themselves, while the reporter activities are resulted from the sequences containing both RUNX1 and GATA sites. Thus, these data indicate that the capacity of transactivation of functional elements is not necessarily consistent with their in vitro binding capacity. In addition, the in vitro binding capacity does not, as most likely that often reflect. A similar result was observed with nuclear extract from Jurkat cells with a slightly different pattern of formed DNA-protein complex, which was specific as judged by the supershifted band (Fig. 4). In contrast to RUNX1 binding, a DNA-protein complex was observed but not supershifted by the two anti-PU.1 antibodies used (Fig. 4).
In summary, we for the first time demonstrate the involvement of RUNX1 and PU.1 in transcriptional control of an eosinophil-specific gene. Thus, RUNX1 and PU.1 might regulate the CCR3 gene by binding their cis-acting elements in its regulatory region in conjunction with GATA-1.
Figures and Tables
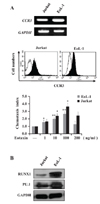 | Figure 1Expression of CCR3, RUNX1, and PU.1 in Jurkat and EoL-1 cells. (A) Expression of CCR3 mRNA and protein was analyzed using RT-PCR and FACS, respectively, and chemotactic responses to eotaxin (1~200 ng/ml) was analyzed. The results shown are representative of three to five independent experiments. Statistical significance was obtained using SPSS, compared with medium. *p<0.05 and **p<0.01. (B) Expression of RUNX1 and PU.1 proteins was analyzed using Western blotting. GAPDH mRNA and protein were used as loading controls. The results shown are representative of three independent experiments. |
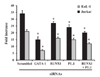 | Figure 2Effects of RUNX1 and PU.1 siRNAs on CCR3 reporter activity. Cells were co-transfected with 50 nM siRNAs for RUNX1, PU.1, GATA-1 or scramble along with the CCR3 reporter. After 36 h, luciferase activity was measured and normalized to Renilla luciferase activity. Data represent the mean±SEM of three independent experiments. Statistical significance was obtained using ANOVA, compared with scrambled siRNA. *p<0.01. |
 | Figure 3Effects of point mutants of RUNX1 and PU.1 binding motifs on CCR3 reporter activity. (A) Nucleotide sequence of the regulatory region construct 1 containing exon 1 and proximal intron 1 of the CCR3 gene. The sites for RUNX1 and PU.1 are underlined, as is the functional GATA site. (B) Point mutants were constructed and transfected into EoL-1 and Jurkat cells. Reporter activity was measured 48 h post-transfection. Transfection efficiency was normalized to cotransfected Renilla luciferase activity. Open and filled symbols indicate wild-type and mutant sites, respectively. Data represent the mean±SEM of three independent experiments performed in triplicate. *p<0.01. |
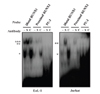 | Figure 4Binding of RUNX1 and PU.1 to their putative sites on the CCR3 regulatory region. Nuclear extracts of EoL-1 and Jurkat cells were incubated with probes harboring RUNX1 and PU.1 binding elements in the presence or absence of their respective Abs. S and C indicate specific Ab and control Ab, respectively. *, **, and *** indicate specific binding, a supershift, and non-specific binding, respectively. |
Table I
Primers used in overlap extension PCR to generate point mutants

ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2013R1A1A2058266 to IYC).
References
1. Pease JE, Williams TJ. Chemokines and their receptors in allergic disease. J Allergy Clin Immunol. 2006; 118:305–318.

3. Willems LI, Ijzerman AP. Small molecule antagonists for chemokine CCR3 receptors. Med Res Rev. 2010; 30:778–817.

4. Mori Y, Iwasaki H, Kohno K, Yoshimoto G, Kikushige Y, Okeda A, Uike N, Niiro H, Takenaka K, Nagafuji K, Miyamoto T, Harada M, Takatsu K, Akashi K. Identification of the human eosinophil lineage-committed progenitor: revision of phenotypic definition of the human common myeloid progenitor. J Exp Med. 2009; 206:183–193.

5. Voehringer D, van RN, Locksley RM. Eosinophils develop in distinct stages and are recruited to peripheral sites by alternatively activated macrophages. J Leukoc Biol. 2007; 81:1434–1444.

6. Kim BS, Uhm TG, Lee SK, Lee SH, Kang JH, Park CS, Chung IY. The crucial role of GATA-1 in CCR3 gene transcription: modulated balance by multiple GATA elements in the CCR3 regulatory region. J Immunol. 2010; 185:6866–6875.

7. Vijh S, Dayhoff DE, Wang CE, Imam Z, Ehrenberg PK, Michael NL. Transcription regulation of human chemokine receptor CCR3: evidence for a rare TATA-less promoter structure conserved between drosophila and humans. Genomics. 2002; 80:86–95.

8. Zimmermann N, Daugherty BL, Kavanaugh JL, El-Awar FY, Moulton EA, Rothenberg ME. Analysis of the CC chemokine receptor 3 gene reveals a complex 5' exon organization, a functional role for untranslated exon 1, and a broadly active promoter with eosinophil-selective elements. Blood. 2000; 96:2346–2354.

9. Scotet E, Schroeder S, Lanzavecchia A. Molecular regulation of CC-chemokine receptor 3 expression in human T helper 2 cells. Blood. 2001; 98:2568–2570.

10. Klemsz MJ, McKercher SR, Celada A, Van BC, Maki RA. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990; 61:113–124.

11. Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994; 265:1573–1577.

12. McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, Maki RA. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996; 15:5647–5658.

13. DeKoter RP, Walsh JC, Singh H. PU.1 regulates both cytokine-dependent proliferation and differentiation of granulocyte/macrophage progenitors. EMBO J. 1998; 17:4456–4468.

14. McNagny KM, Sieweke MH, Doderlein G, Graf T, Nerlov C. Regulation of eosinophil-specific gene expression by a C/EBP-Ets complex and GATA-1. EMBO J. 1998; 17:3669–3680.

15. Kodandapani R, Pio F, Ni CZ, Piccialli G, Klemsz M, McKercher S, Maki RA, Ely KR. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETSdomain-DNA complex. Nature. 1996; 380:456–460.

16. Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, Curley DP, Kutok JL, Akashi K, Williams IR, Speck NA, Gilliland DG. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005; 106:494–504.

17. Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009; 9:106–115.

18. Mukai K, BenBarak MJ, Tachibana M, Nishida K, Karasuyama H, Taniuchi I, Galli SJ. Critical role of P1-Runx1 in mouse basophil development. Blood. 2012; 120:76–85.

19. Meyers S, Downing JR, Hiebert SW. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol. 1993; 13:6336–6345.

20. Kong SK, Kim BS, Uhm TG, Lee W, Lee GR, Park CS, Lee CH, Chung IY. Different GATA factors dictate CCR3 transcription in allergic inflammatory cells in a cell type-specific manner. J Immunol. 2013; 190:5747–5756.

21. Elagib KE, Racke FK, Mogass M, Khetawat R, Delehanty LL, Goldfarb AN. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003; 101:4333–4341.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download