Abstract
Purification of enough numbers of circulating eosinophils is difficult because eosinophils account for less than 5% peripheral blood leukocytes. Human eosinophilic leukemia EoL-1 cells have been considered an in vitro source of eosinophils as they can differentiate into mature eosinophil-like cells when incubated with dibutyryl cAMP (dbcAMP) or butyric acid. In this study, the viability and phenotypic maturation of EoL-1 cells stimulated by either dbcAMP or butyric acid were comparatively analyzed. After treatment with 100 µM dbcAMP or 0.5 µM butyric acid, EoL-1 cells showed morphological signs of differentiation, although the number of nonviable EoL-1 cells was significantly increased following butyric acid treatment. Stimulation of EoL-1 cells with 0.5 µM butyric acid more effectively induced the expression of mature eosinophil markers than stimulation with dbcAMP. These results suggest that treatment of EoL-1 cells with 0.5 µM butyric acid for limited duration could be an effective strategy for inducing their differentiation. Considering that expression of CCR3 was not sufficient in EoL-1 cells stimulated with 0.5 µM butyric acid, treatment of the chemically stimulated EoL-1 cells with cytokines, which primarily support eosinophil maturation, would help to obtain differentiated EoL-1 cells with greater functional maturity.
Eosinophils are multifunctional leukocytes that have been implicated in the pathogenesis of Th2-type inflammatory processes, including helminth infections and allergic diseases. The cytoplasm of mature eosinophils contains numerous secondary granules such as eosinophil peroxidase, eosinophil cationic protein, eosinophil-derived neurotoxin, and major basic protein, and the exocytotic release of these granule-derived cytotoxic proteins contributes to inflammatory responses induced by eosinophil activation (12). Eosinophils are produced in the bone marrow from pluripotential stem cells, which differentiate into eosinophil progenitors marked by CD34+IL-5Rα+ expression (1). Eosinophil lineage specification is determined by the interplay of several transcription factors, including GATA-1 (a zinc finger family member), PU.1 (an ETS family member), and members of the CCAAT/enhancer-binding protein (C/EBP) family (345). Following differentiation, permissive proliferation and migration of eosinophils from the bone marrow to the circulation are regulated primarily by IL-5. However, eosinophils account for less than 5% peripheral blood leukocytes in normal humans and have a short life span, making it difficult to study the biological properties of circulating eosinophils in vitro (6).
A human eosinophilic cell line, EoL-1, has been considered a useful in vitro model to study human eosinophils (78). EoL-1 cells can be induced to develop into eosinophilic granule-containing cells by chemical stimuli, including dibutyryl cAMP (dbcAMP) and butyric acid (9). There are several markers for mature eosinophils, including CCR3 and IL-5Rα (1011). Although independently performed studies have revealed maturation of EoL-1 cells following stimulation by either dbcAMP or butyric acid, comparative phenotypic analysis of differentiated EoL-1 cells will contribute to suggesting optimal EoL-1 stimulating conditions compatible with experimental purpose. In this study, we found that stimulation with butyric acid was more effective than stimulation with dbcAMP for induction of EoL-1 cell differentiation. However, both butyric acid and dbcAMP were not sufficient for the expression of CCR3 in EoL-1 cells, and we propose subsequent cytokine treatment of chemically stimulated EoL-1 cells.
EoL-1 cells (DSMZ, Braunschweig, Germany) were maintained in RPMI 1640 medium (Sigma, St. Louis, MO, USA) supplemented with 10% FBS (Gibco Laboratories, Grand Island, NY, USA) in 5% CO2 at 37℃. EoL-1 cells were induced to differentiate by the addition of dbcAMP (Sigma-Aldrich) or butyric acid (Sigma-Aldrich) for 9 days. The cell concentration was adjusted to 5×105/ml every 3 days.
For morphological analysis, cultured EoL-1 cells were spun at 500 rpm for 5 min on glass slides (Cytospin 3, Shandon, Pittsburgh, PA, USA). The slides were air-dried, stained with Diff-Quik stain solution (Sysmex, Kobe, Japan) and observed using the CX41 microscope (Olympus, Tokyo, Japan).
Aliquots of EoL-1 cells, before and after stimulation with dbcAMP or butyric acid, were resuspended in FACS buffer (PBS containing 10% FBS, 10 mM EDTA, 20 mM HEPES, and 1 mM sodium pyruvate). After Fc receptor blocking with human TruStain FcX (Biolegend, San Diego, CA, USA) for 15 min at 4℃, the cells were stained with antibodies against FITC anti-human CCR3 (R&D, Minneapolis, MN, USA) or PE anti-human IL5Rα (R&D). For analysis of viability, cells were incubated with 7-amino-actinomycin D (Biolegend) for 10 min at room temperature. Each sample was analyzed using FACSCalibur (BD Bioscience, San Diego, CA, USA) and the data were processed with FlowJo software (Tree Star, Ashland, OR, USA).
RNA from unstimulated and dbcAMP or butyric acid treated EoL-1 cells was extracted using QIAzol lysis reagent (Qiagen, Hilden, Germany) and was column-purified with an RNeasy Mini Kit (Qiagen). The purified RNA (500 ng) was treated with DNase I (New England Biolabs, Ipswich, MA, USA), and cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). Real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad). PCR was performed using a CFX96 Real-Time System (Bio-Rad). Sequences of primers are shown in Table S1.
Treatment of EoL-1 cells with either 100 µM dbcAMP or 0.5 µM butyric acid for 6 to 9 days is known to induce eosinophilic maturation (91213). EoL-1 cells represent undifferentiated promyelocytic eosinophils possessing a large nucleus with prominent nucleoli (Fig. 1A) (14). After treatment with dbcAMP or butyric acid, we observed morphological signs of EoL-1 differentiation, including nuclear lobulation and increased proportion of cytoplasm to nucleus (Fig. 1A). However, compared to the group stimulated with 100 µM dbcAMP, EoL-1 cells stimulated with 0.5 µM butyric acid showed remarkable increase in apoptotic populations exhibiting cellular shrinkage and nucleus condensation (Fig. 1A). Consistent with the morphological findings, stimulation with 0.5 µM butyric acid significantly inhibited the proliferation of EoL-1 cells after 2 days (Fig. 1B). The proliferation of EoL-1 cells treated with dbcAMP (100 µM and 10 µM) or 0.05 µM butyric acid increased in a time-dependent manner during 8 days of incubation (Fig. 1B). Additionally, compared to that in other groups, the number of 7-amino-actinomycin D-positive nonviable cells remarkably increased in the group stimulated with 0.5 µM butyric acid (Fig. 1C). These data indicate that stimulation of EoL-1 cells with 0.5 µM butyric acid for prolonged period is unsuitable for EoL-1 cells in vitro assay in terms of cell viability.
Next, we examined the expression of PRG2, EPX, CCR3, IL5RA, and GATA1, markers for mature eosinophils, in EoL-1 cells stimulated with dbcAMP or butyric acid. As shown in Fig. 2A, 0.5 µM butyric acid effectively induced the expression of PRG2, EPX, CCR3, IL5RA, and GATA1 in EoL-1 cells than 100 µM dbcAMP in a time-dependent manner. Additionally, the effect of 100 µM dbcAMP stimulation was limited to the expression of PRG2 and EPX, which encode cytoplasmic granules of eosinophils (Fig. 2B). However, it could be plausible that CCR3 levels could be low even in the 0.5 µM butyric acid-treated group, considering the relatively low fold increase of the transcript in these cells (Fig. 2). Consistent with this idea, mean fluorescence intensity (MFI) of CCR3 expression was not significantly higher in 0.5 µM butyric acid-treated EoL-1 cells than in unstimulated control cells (Fig. 3). Technical problems related to reagents used for flow cytometry analysis were excluded by demonstrating relatively robust expression of CCR3 and IL-5Rα in eosinophils obtained from human peripheral blood (MFI=145±5, MFI=309±11, respectively, Fig. S1). Collectively, these data indicate that compared to dbcAMP treatment, treatment with 0.5 µM butyric acid was more effective in inducing phenotypic maturation of EoL-1 cells, and that the treatment duration should be less than 5 days to preserve the viability of the stimulated cells.
Eosinophil lineage-committed progenitors developed in the bone marrow are identified via surface expression of IL-5Rα and mature into eosinophil precursors containing cytoplasmic granules (15). Eosinophil precursors contain a granule-rich cytoplasm, and their permissive proliferation and differentiation into mature eosinophils is regulated by several cytokines, including IL-5, IL-3, and GM-CSF (16). Recently it was reported that eosinophil lineage-committed progenitors or eosinophil precursors in the mouse bone marrow do not express CCR3 (17). IL-5 primarily induces maturation of eosinophils and stimulates eosinophil migration out of the bone marrow to the circulation mediated by eosinophils expressing CCR3. We have also reported the expression of CCR3 in EoL-1 cells treated with IL-3 and GM-CSF following stimulation with dbcAMP (6). Therefore, we suggest that additional cytokine treatment would be needed to induce differentiation of EoL-1 cells into functionally mature phenotype following chemical stimulation.
Figures and Tables
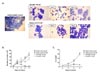 | Figure 1Effects of dbcAMP and butyric acid on the morphologic features and proliferation capacity of EoL-1 cells. (A) EoL-1 cells were incubated for the indicated periods in the absence or presence of 100 µM dbcAMP or 0.5 µM butyric acid. Cell number was adjusted to 5×105/ml every 3 days. Diff-Quik staining of unstimulated EoL-1 cells and EoL-1 cells stimulated with dbcAMP or butyric acid. Arrows denote cells showing nuclear lobulation and arrow heads indicate cells showing shrinkage and chromatin condensation. Original magnification, ×40. (B, C) EoL-1 cells were incubated for 8 or 9 days in medium containing indicated concentration of dbcAMP or butyric acid. The cells were then harvested and enumerated (B) and the viability of the cells was determined by flow cytometry analysis of 7-amino-actinomycin D (7-AAD) (C). All data are representative of two or more independent experiments. Data are mean±s.e.m. values. *p<0.05, **p<0.01, and ***p<0.001 (Student's t-test) vs. the control. |
 | Figure 2Effects of dbcAMP and butyric acid on the differentiation of EoL-1 cells. (A) cDNA was prepared from total RNA obtained from undifferentiated EoL-1 cells (day 0) and EoL-1 cells stimulated with indicated concentrations dbcAMP or butyric acid for 3 or 6 days. mRNA expressions of PRG2, EPX, CCR3, Il5RA, and GATA1 were analyzed by real-time PCR. Data are mean±s.e.m. values. *p<0.05, **p<0.01, and ***p<0.001 (Student's t-test) of 0.5 µM butyric acid vs. 100 µM dbcAMP stimulation. (B) mRNA expressions of PRG2, EPX, CCR3, Il5RA, and GATA1 of undifferentiated EoL-1 cells (control) and EoL-1 cells stimulated with 100 µM dbcAMP or 0.5 µM butyric acid for 3 or 6 days. Data are mean±s.e.m. values. *p<0.05, **p<0.01, and ***p<0.001 (Student's t-test) vs. the control. |
 | Figure 3Effects of dbcAMP and butyric acid on the expression of CCR3 and IL-5Rα in EoL-1 cells. The expression of CCR3 or IL-5Rα in undifferentiated EoL-1 cells and EoL-1 cells stimulated with 100 µM dbcAMP or 0.5 µM butyric acid for 6 days was determined by flow cytometry. Flow cytometric expression of CCR3 or IL-5Rα in indicated cell groups was shown as mean fluorescence intensity (MFI). Data are representative of two or more independent experiments. Data are mean±s.e.m. values. **p<0.01 (Student's t-test) vs. the control. |
ACKNOWLEDGEMENTS
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2013-R1A1A2004820). The author thanks to Ye-Rang Chung (Gachon University, Korea) for technical assistance.
References
2. Rothenberg ME. Eosinophilic gastrointestinal disorders (EGID). J Allergy Clin Immunol. 2004; 113:11–28.

3. Milanovic M, Terszowski G, Struck D, Liesenfeld O, Carstanjen D. IFN consensus sequence binding protein (Icsbp) is critical for eosinophil development. J Immunol. 2008; 181:5045–5053.

4. Hogan SP, Waddell A, Fulkerson PC. Eosinophils in infection and intestinal immunity. Curr Opin Gastroenterol. 2013; 29:7–14.

5. Jung Y, Rothenberg ME. Roles and regulation of gastrointestinal eosinophils in immunity and disease. J Immunol. 2014; 193:999–1005.

6. Jung YJ, Woo SY, Jang MH, Miyasaka M, Ryu KH, Park HK, Seoh JY. Human eosinophils show chemotaxis to lymphoid chemokines and exhibit antigen-presenting-cell-like properties upon stimulation with IFN-gamma, IL-3 and GM-CSF. Int Arch Allergy Immunol. 2008; 146:227–234.

7. Saito H, Bourinbaiar A, Ginsburg M, Minato K, Ceresi E, Yamada K, Machover D, Breard J, Mathe G. Establishment and characterization of a new human eosinophilic leukemia cell line. Blood. 1985; 66:1233–1240.

9. Wong CK, Ho CY, Lam CW, Zhang JP, Hjelm NM. Differentiation of a human eosinophilic leukemic cell line, EoL-1: characterization by the expression of cytokine receptors, adhesion molecules, CD95 and eosinophilic cationic protein (ECP). Immunol Lett. 1999; 68:317–323.

10. Yoshida T, Ikuta K, Sugaya H, Maki K, Takagi M, Kanazawa H, Sunaga S, Kinashi T, Yoshimura K, Miyazaki J, Takaki S, Takatsu K. Defective B-1 cell development and impaired immunity against Angiostrongylus cantonensis in IL-5R alpha-deficient mice. Immunity. 1996; 4:483–494.

11. Kitaura M, Nakajima T, Imai T, Harada S, Combadiere C, Tiffany HL, Murphy PM, Yoshie O. Molecular cloning of human eotaxin, an eosinophil-selective CC chemokine, and identification of a specific eosinophil eotaxin receptor, CC chemokine receptor 3. J Biol Chem. 1996; 271:7725–7730.

12. Ishihara K, Takahashi A, Kaneko M, Sugeno H, Hirasawa N, Hong J, Zee O, Ohuchi K. Differentiation of eosinophilic leukemia EoL-1 cells into eosinophils induced by histone deacetylase inhibitors. Life Sci. 2007; 80:1213–1220.

13. Saito H, Hayakawa T, Mita H, Akiyama K, Shida T. Effect of butyric acid on induction of differentiation into eosinophil-like cells in human eosinophilic leukemia cells, EoL-1 cell line: possible role of granulocyte-macrophage colony-stimulating factor as an autocrine differentiating factor. Int Arch Allergy Immunol. 1993; 100:240–247.

14. Morita M, Saito H, Honjo T, Saito Y, Tsuruta S, Kim KM, Tanaka M, Mori KJ, Mayumi M, Mikawa H. Differentiation of a human eosinophilic leukemia cell line (EoL-1) by a human T-cell leukemia cell line (HIL-3)-derived factor. Blood. 1991; 77:1766–1775.

15. Iwasaki H, Mizuno S, Mayfield R, Shigematsu H, Arinobu Y, Seed B, Gurish MF, Takatsu K, Akashi K. Identification of eosinophil lineage-committed progenitors in the murine bone marrow. J Exp Med. 2005; 201:1891–1897.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download