1. Roopenian D, Choi EY, Brown A. The immunogenomics of minor histocompatibility antigens. Immunol Rev. 2002; 190:86–94. PMID:
12493008.

2. Choi EY, Yoshimura Y, Christianson GJ, Sproule TJ, Malarkannan S, Shastri N, Joyce S, Roopenian DC. Quantitative analysis of the immune response to mouse non-MHC transplantation antigens
in vivo: the H60 histocompatibility antigen dominates over all others. J Immunol. 2001; 166:4370–4379. PMID:
11254691.
3. Choi EY, Christianson GJ, Yoshimura Y, Jung N, Sproule TJ, Malarkannan S, Joyce S, Roopenian DC. Real-time T-cell profiling identifies H60 as a major minor histocompatibility antigen in murine graft-versus-host disease. Blood. 2002; 100:4259–4265. PMID:
12393464.

4. Choi JH, Ryu SJ, Jung KM, Kim S, Chang J, Kim TW, Choi EY. TCR diversity of H60-specific CD8 T cells during the response evolution and influence of CD4 help. Transplantation. 2009; 87:1609–1616. PMID:
19502951.

5. Ryu SJ, Jung KM, Yoo HS, Kim TW, Kim S, Chang J, Choi EY. Cognate CD4 help is essential for the reactivation and expansion of CD8 memory T cells directed against the hematopoietic cell-specific dominant minor histocompatibility antigen, H60. Blood. 2009; 113:4273–4280. PMID:
19139082.

6. Choi EY, Christianson GJ, Yoshimura Y, Sproule TJ, Jung N, Joyce S, Roopenian DC. Immunodominance of H60 is caused by an abnormally high precursor T cell pool directed against its unique minor histocompatibility antigen peptide. Immunity. 2002; 17:593–603. PMID:
12433366.

7. Malarkannan S, Shih PP, Eden PA, Horng T, Zuberi AR, Christianson G, Roopenian D, Shastri N. The molecular and functional characterization of a dominant minor H antigen, H60. J Immunol. 1998; 161:3501–3509. PMID:
9759870.
8. Malarkannan S, Horng T, Eden P, Gonzalez F, Shih P, Brouwenstijn N, Klinge H, Christianson G, Roopenian D, Shastri N. Differences that matter: major cytotoxic T cell-stimulating minor histocompatibility antigens. Immunity. 2000; 13:333–344. PMID:
11021531.
9. Jeon JY, Jung KM, Chang J, Choi EY. Characterization of CTL Clones Specific for Single Antigen, H60 Minor Histocompatibility Antigen. Immune Netw. 2011; 11:100–106. PMID:
21637387.

10. Russell PS, Chase CM, Madsen JC, Hirohashi T, Cornell LD, Sproule TJ, Colvin RB, Roopenian DC. Coronary artery disease from isolated non-H2-determined incompatibilities in transplanted mouse hearts. Transplantation. 2011; 91:847–852. PMID:
21378606.

11. Ryu SJ, Jeon JY, Chang J, Sproule TJ, Roopenian DC, Choi EY. A single-amino-acid variant of the H60 CD8 epitope generates specific immunity with diverse TCR recruitment. Mol Cells. 2012; 33:393–399. PMID:
22441676.

12. Kouskoff V, Signorelli K, Benoist C, Mathis D. Cassette vectors directing expression of T cell receptor genes in transgenic mice. J Immunol Methods. 1995; 180:273–280. PMID:
7714342.

13. Kim J, Ryu SJ, Oh K, Ju JM, Jeon JY, Nam G, Lee DS, Kim HR, Young Kim J, Chang J, Sproule T, Choi K, Roopenian D, Young CE. Memory programming in CD8(+) T-cell differentiation is intrinsic and is not determined by CD4 help. Nat Commun. 2015; 6:7994. PMID:
26272364.

14. Ehst BD, Ingulli E, Jenkins MK. Development of a novel transgenic mouse for the study of interactions between CD4 and CD8 T cells during graft rejection. Am J Transplant. 2003; 3:1355–1362. PMID:
14525595.

15. Barthlott T, Stockinger B. Lineage fate alteration of thymocytes developing in an MHC environment containing MHC/peptide ligands with antagonist properties. Eur J Immunol. 2001; 31:3595–3601. PMID:
11745379.
16. Williams O, Tarazona R, Wack A, Harker N, Roderick K, Kioussis D. Interactions with multiple peptide ligands determine the fate of developing thymocytes. Proc Natl Acad Sci U S A. 1998; 95:5706–5711. PMID:
9576948.

17. Speir JA, Stevens J, Joly E, Butcher GW, Wilson IA. Two different, highly exposed, bulged structures for an unusually long peptide bound to rat MHC class I RT1-Aa. Immunity. 2001; 14:81–92. PMID:
11163232.

18. Apostolopoulos V, Yuriev E, Ramsland PA, Halton J, Osinski C, Li W, Plebanski M, Paulsen H, McKenzie IF. A glycopeptide in complex with MHC class I uses the GalNAc residue as an anchor. Proc Natl Acad Sci U S A. 2003; 100:15029–15034. PMID:
14657390.

19. Boesteanu A, Brehm M, Mylin LM, Christianson GJ, Tevethia SS, Roopenian DC, Joyce S. A molecular basis for how a single TCR interfaces multiple ligands. J Immunol. 1998; 161:4719–4727. PMID:
9794402.
20. Anderson AC, Nicholson LB, Legge KL, Turchin V, Zaghouani H, Kuchroo VK. High frequency of autoreactive myelin proteolipid protein-specific T cells in the periphery of naive mice: mechanisms of selection of the self-reactive repertoire. J Exp Med. 2000; 191:761–770. PMID:
10704458.
21. Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire:what thymocytes see (and don't see). Nat Rev Immunol. 2014; 14:377–391. PMID:
24830344.
22. Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003; 21:139–176. PMID:
12414722.

23. Sprent J, Kishimoto H. The thymus and negative selection. Immunol Rev. 2002; 185:126–135. PMID:
12190927.

24. Modigliani Y, Thomas-Vaslin V, Bandeira A, Coltey M, Le Douarin NM, Coutinho A, Salaun J. Lymphocytes selected in allogeneic thymic epithelium mediate dominant tolerance toward tissue grafts of the thymic epithelium haplotype. Proc Natl Acad Sci USA. 1995; 92:7555–7559. PMID:
7638230.

25. Mayerova D, Hogquist KA. Central tolerance to self-antigen expressed by cortical epithelial cells. J Immunol. 2004; 172:851–856. PMID:
14707055.

26. Goldman KP, Park CS, Kim M, Matzinger P, Anderson CC. Thymic cortical epithelium induces self tolerance. Eur J Immunol. 2005; 35:709–717. PMID:
15719367.

27. McCaughtry TM, Baldwin TA, Wilken MS, Hogquist KA. Clonal deletion of thymocytes can occur in the cortex with no involvement of the medulla. J Exp Med. 2008; 205:2575–2584. PMID:
18936237.

28. Baldwin TA, Sandau MM, Jameson SC, Hogquist KA. The timing of TCR alpha expression critically influences T cell development and selection. J Exp Med. 2005; 20:111–121. PMID:
15998791.
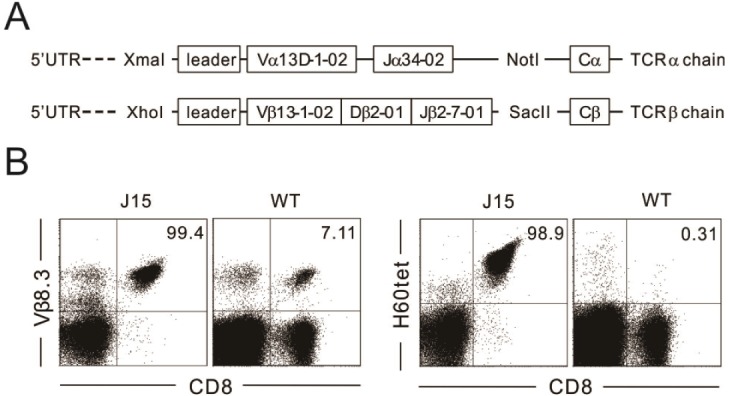

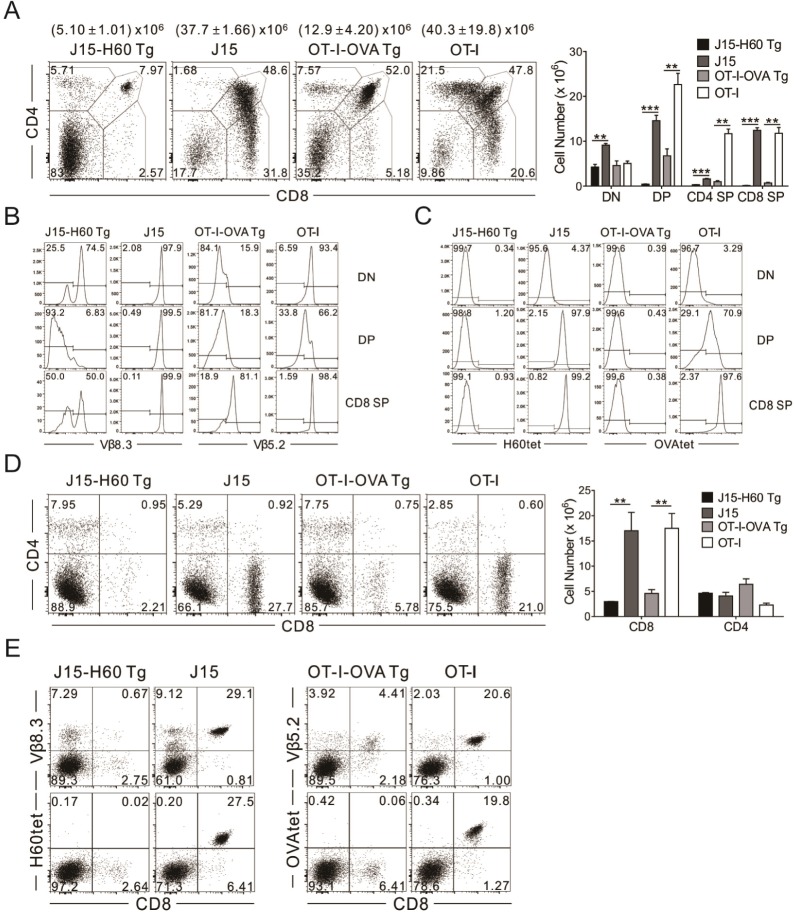





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download