Abstract
T cells are central players in the regulation of adaptive immunity and immune tolerance. In the periphery, T cell differentiation for maturation and effector function is regulated by a number of factors. Various factors such as antigens, co-stimulation signals, and cytokines regulate T cell differentiation into functionally specialized effector and regulatory T cells. Other factors such as nutrients, micronutrients, nuclear hormones and microbial products provide important environmental cues for T cell differentiation. A mounting body of evidence indicates that the microbial metabolites short-chain fatty acids (SCFAs) have profound effects on T cells and directly and indirectly regulate their differentiation. We review the current status of our understanding of SCFA functions in regulation of peripheral T cell activity and discuss their impact on tissue inflammation.
Commensal microbiota functions not only to serve as targets of host immunity but also as active players in regulation of host physiology and immunity as a result of long-term coevolution of the host and microbes. T cells play central roles in the regulation of anti-microbial immunity and tissue inflammation. Most major T cell types are made in the thymus, although extrathymic generation of some T cell subsets has been described (1,2). T cells are divided into the major TCR-αβ and minor γδ T cell groups. αβ T cells are highly heterogeneous and grouped into CD4+ conventional T cells, CD8+ conventional T cells, NKT cells, and other innate TCRα-expressing T cells such as mucosal-associated invariant T (MAIT) cells (3,4,5,6). CD4+ conventional T cells are further divided into FoxP3+ regulatory and FoxP3- T cells (7,8). FoxP3-CD4+ T cells include various effector and regulatory T cells based on their cytokine phenotype (IFNγ, IL-17, IL-22, IL-4, IL-9, IL-10, IL-35, and/or LAP-TGFβ1) (6,9). These T helper cells include IFNγ+ Th1 cells, IL-17/IL-22+ Th17 cells, IL-4+ Th2 cells, IL-9+ Th9 cells, IL-21+ T-FH cells, and IL-10/IL-35/TGFβ1+ Tregs (9,10,11,12). All of these T helper cell subsets are generated mainly in the periphery from naïve T cells made in the thymus. TCR repertoire and antigen specificity/affinity greatly influence T cell differentiation in the thymus and periphery (13,14). Co-stimulation signals such as CD28, ICOS, CTLA4, OX-40, and PD-1 signaling reciprocally regulate T cell differentiation and effector function (15,16,17). Cytokine milieu during T cell activation is crucial to generate specialized effector versus regulatory T cell subsets (6,9). A mounting body of evidence indicates that nutrients and metabolites provide significant regulatory signals for T cell differentiation (18,19,20,21,22,23). Potentially important roles of gut microbial products such as short-chain fatty acids (SCFAs) have been recently documented (24,25,26). In this review, we will review the recent progress in our understanding of the roles of SCFAs in regulating CD4+ T helper cell differentiation and the impact of this process on tissue inflammation.
SCFAs refer to free fatty acids containing fewer than 6 carbons and therefore they have short aliphatic carbon-chains. Formic acid (C1), acetic acid (C2), propionic acid (C3), butyric acid (C4), and valeric acid (C5) belong to the SCFA group. These metabolites are distinguished from longer fatty acids such as medium-chain (6-12 carbons) and long-chain free fatty acids. Because they have relatively shorter hydrophobic chains as well as the hydrophilic carboxyl group, SCFAs are water soluble and readily absorbed or transported into cells. SCFAs are produced by gut microbiota as fermentation products, meaning that they are partially oxidized from sugar molecules under anaerobic conditions in the colon. Carbohydrates are good sources of SCFAs but SCFAs can be made from other nutrients such as proteins and peptides albeit at low levels (27). These SCFA precursors, however, are easily degraded by host digestive enzymes in the upper alimentary tract and don't reach the microbiota in the colon in significant amounts for SCFA production. In contrast, digestion-resistant oligosaccharides and fibers (e.g. oligofructose, inulin, pectin, and arabinoxylan) are good sources of SCFAs. Insoluble fibers including cellulose and chitin, however, are not readily fermented by the microbiota and thus do not produce SCFAs at significant levels.
While it is yet to be determined clearly through extensive bacterial isolation and metagenomics studies, available information indicates that bacteria species greatly differ in their genetic make-up of enzymes involved in SCFA production (28,29). Among SCFAs, C2 is relatively more readily produced than C3 and C4 by most enteric and acetogenic bacteria (30). Propionate can be produced by three pathways (i.e. succinate, acrylate, and propanediol) from various sugar molecules such as pentoses, hexoses, and rhamnose (31). Bacteroidetes and some Firmicutes are good producers of C3 mainly through the succinate pathway. Production of C4 requires additional enzymatic processes that extend acetyl-CoA with butyryl-CoA:acetate CoA-transferase, which is active in some bacteria including Roseburia, Eubacterium and Anaerostipes species and Faecalibacterium prausnitzii (27,32).
The combined concentrations of SCFAs produced in the colon reach ~150 mM, making SCFAs the most abundant anions in the colon. SCFAs are absorbed in the colon and either utilized in colonocytes or transported via the portal vein to reach the blood circulation and other organs. The liver and muscle are major systemic organs for SCFA metabolism and consumption. SCFAs enter cells through passive diffusion and carrier-mediated transportation through SMCT1/SLC5a8 and MCT1/SLC16a1 (33,34,35). SMCT1 is a sodium-coupled monocarboxylate transporter 1 for cell intake of SCFAs and related organic acids such as lactate and pyruvate (34). SMCT1 belongs to the SLC5 Na+/glucose cotransporter gene family (33). MCT1 is an H+-coupled transporter for SCFAs and related organic acids and it transports these molecules depending on the net chemical gradients for H+ and monocarboxylates across the membrane (36). Expression of these transporters in the apical membrane of colonocytes, DCs, kidney cells, and/or brain cells has been documented (Table I).
SCFAs activate several G-protein-coupled cell surface receptors (GPCR). GPR41 and GPR43 are major receptors that can be activated by most SCFAs (37). Gut enteroendocrine cells highly express GPR41 and GPR43 (38,39). Other regular enterocytes express these receptors also at functional levels (38,39,40,41). GPR41 is also expressed in adipocytes, renal smooth muscle cells, enteric neuronal cells, and pancreatic cells (Table I) (42,43). The expression of GPR41 is co-regulated with GPR40, a receptor for medium and long-chain fatty acids, because their gene transcription is regulated by the same promoter (44). GPR43 is expressed by granulocytes and some myeloid cells (45,46,47). GPR109a, a receptor for niacin (also called nicotinic acid and vitamin B3), is a receptor also for C4 (48). GPR109a is expressed by gut epithelial cells, adipocytes, macrophages and dendritic cells (Table I). Olfr78 is expressed in the kidney juxtaglomerular apparatus and is activated by C2 and C3 (49). However, T cells do not express these receptors at functionally significant levels (unpublished results) (24). Major cell types expressing these receptors are listed in Table I.
SCFAs, also called volatile fatty acids because of their relatively more volatile nature compared to longer fatty acids, have been studied for more than a century (50,51). These early observations linked SCFAs to diarrhea and ion balance in the intestine. SCFAs are physiologically important in the intestine as they regulate ion absorption and gut motility. Because SCFAs are absorbed first into colonic epithelial cells and can be metabolized in these cells, they profoundly affect the basic biology of intestinal epithelial cells. SCFAs, particularly C4, are used as the major energy source for colonic epithelial cells and regulate their gene expression, proliferation, differentiation, and apoptosis (52). For example, SCFAs promote the production of mucin and gastrointestinal peptide (e.g. LL-37) (53), molecules important for gut barrier function.
SCFAs condition intestinal epithelial cells to make them more readily respond to bacterial products (40). This function is important to prepare epithelial cells for mounting optimal innate immune responses to invading pathogens and commensal bacteria, and therefore helps prevent chronic intestinal inflammatory responses to microbes and their products. In this regard, SCFAs have anti-inflammatory activity in regulating intestinal inflammation (54). Intestinal epithelial cells express GPR41, GPR43, and GPR109a, which mediate a significant portion of the SCFA function (48,55,56,57). These GPCRs activate signaling processes such as RAS, protein kinase A, PI3K, and ERK1/2 for activation of transcription factors such as ATF2 (40,47,48,58,59). Activation of this pathway is important for expression of key immune and inflammatory mediators such as IL-1, IL-6, TNF-α, CXCL1, and CXCL2. Another function of SCFAs is to activate GPR41 and GPR43 on secretory epithelial cells to produce glucagon-like peptide (GLP)-1 (60). Enteric neurons express GPR41 to sense SCFAs for regulation of gut motility (39). To support this, there is a high correlation in expression sites between SCFA receptors and gut endocrine hormones such as GLP-1, PYY, and neurotensin. Another major mechanism for the SCFA regulation of epithelial cells is mediated through inhibition of HDACs by SCFAs (61,62).
Other cell types are also regulated by SCFAs. SCFAs induce the chemotaxis of neutrophils via activation of GPR43 (58,59) and regulate neutrophil degranulation (63,64). SCFAs also regulate macrophages and dendritic cells (DCs) (65,66). SCFAs suppresses NF-kB and the production of inflammatory cytokines such as IL-6 and TNF-α but increases IL-10 secretion from macrophages (67). In contrast, increased C2 levels in alcoholism can increase the expression of inflammatory cytokines in macrophages and even exacerbate the inflammatory response in the liver (68). Thus, the SCFA function in regulation of immune responses may be altered in pathological conditions. SCFAs increase satiety and reciprocally regulate adipogenesis and lipolysis (69,70,71). Adipocytes express GPR41 and are activated by SCFAs to produce leptin (42,43,72). Olfr78 activation promotes renin production from the kidney to regulate blood pressure (49).
Early work on C4 revealed its regulatory effect on cytokine production by lymphocytes (73,74). C4 had regulatory effects on production of cytokines such as IL-2, IL-4, IL-5, IL-6, and IL-10 (75). Others observed that C4 induced Fas-upregulation and apoptosis in T cells (76). Smith et al. reported that mice fed with SCFAs had increased numbers of IL-10-producing FoxP3+ T cells in the colon (25). The effect was specific for colonic FoxP3+ T cells, and FoxP3+ T cells in other organs were not expanded after SCFA administration. A mechanism provided by this group for the expanded colonic FoxP3+ T cells was decreased HDAC expression and activity by SCFAs in a GPR43-depednent manner. SCFAs can enter cells through diffusion or carrier-mediated transport and thus do not necessarily go through cell surface receptors. Moreover, T cells do not express GPR43 at significant levels and thus this mechanism remains to be verified. Another group reported that C2 and C3 can directly suppress HDACs and increase histone acetylation at the FoxP3 gene locus for increased transcription (20). Similarly, it was reported that Treg generation was increased by SCFAs as a result of HDAC inhibition by SCFAs and histone H3 acetylation in key regulatory regions of the Foxp3 locus (77). In relation to these reports, inoculation of germ-free mice with SCFA-producing Clostridia groups induced IL-10 and ICOS-expressing FoxP3+ T cells (78). Overall, these studies suggest that SCFAs expand colonic Tregs for immune tolerance.
Our group found that SCFAs can increase IL-10, but not necessarily FoxP3, expression in T cells (24). Interestingly, SCFA either positively or negatively regulate induced FoxP3+ cells depending on the strength of T cell activation in vitro. In high T cell activation conditions, SCFAs can even suppress FoxP3+ cell induction promoted by TGFβ1 and T cell activation. In contrast, SCFAs enhance FoxP3+ cell induction at low T cell activation conditions. Independent of FoxP3 regulation, SCFAs increased IL-10 production in all T cell activation conditions (24). We observed that the FoxP3+ T cells even in the colon were not reproducibly regulated by SCFA administration in vivo (unpublished results). These results imply that FoxP3 induction by SCFAs may be regulated by indirect mechanisms through non-T cells. A rather surprising finding was that SCFAs facilitated naïve T cell differentiation into Th1 and Th17 cells in appropriate T cell polarization conditions. Thus, SCFAs can enhance both effector and regulatory T cells depending on the immunological milieu. In support of this, C2 administration via drinking water increased Th1 and Th17 cells in the intestine and secondary lymphoid tissues during C. rodentium infection (40). In the absence of infection, SCFAs increased gut IL-10+ T cells in vivo, which would promote immune tolerance. It appears that SCFAs selectively promote only the right types of T cells required to handle specific immunological conditions (Fig. 1).
Cells can be regulated by SCFAs largely in three different mechanisms (Fig. 1). The first mechanism involves the activation of SCFA-binding G-protein-coupled receptors (GPCRs) such as GPR41, GPR43, GPR109A, and Olfr78. GPCR signaling can regulate cell activation, proliferation, and differentiation. However, T cells do not express any of these receptors at significant levels, according to published information and unpublished microarray data. Thus, SCFA receptors are not likely to be important for direct regulation of T cells by SCFAs. Another pathway is to regulate cell energy status and relevant signaling processes through integration of SCFAs into cellular metabolism. SCFAs can be converted to Acetyl-CoA and integrated into the citric acid cycle (Krebs cycle). Acetyl-CoA is a central molecule that stores energy in the molecule, which is eventually oxidized to CO2 for energy production. As the result, the cellular energy [ATP/ADP] level increases, and this change boosts mTOR activation (79). In T cells, activation of mTOR skews T cell differentiation into effector T cells such as Th1 and Th17 cells at the expense of FoxP3+ T cells (80). mTOR activation also promotes the generation of IL-10+ cells (81). Thus, the SCFA-regulation of cell metabolism and mTOR accounts for the increased generation of Th1, Th17 cells, and IL-10+ cells. The third mechanism is mediated through the HDAC inhibitor activity of SCFAs (Fig. 1). All major SCFAs such as C2, C3, C4 and C5 have HDAC inhibitor activity (24,82,83). Some regarded that C2 does not have the HDAC inhibitor activity but it has clear HDAC inhibitor activity at concentrations (~10 mM) higher than C3 and C4 (~1 mM) (24). Moreover, C2 is maintained at relatively high concentrations in blood (~1 mM). This HDAC inhibitor activity requires the transport of SCFAs into cells and enzymatic inhibition of HDACs. Class I/II HDACs are major targets of SCFA inhibition. While SCFAs do not suppress class III HDAC such as Sirt1, down-regulation of Sirt1 expression by SCFAs was reported (84). Thus, SCFAs may affect a broad range of HDACs for their regulatory effects. Because HDAC inhibition increases the acetylation of histone and other proteins, the impact of this activity is far reaching and affecting a number of genes and proteins. Physical interaction between HDACs and S6K has been reported (85), and S6K is a downstream effector molecule of the mTOR pathway. P70-S6 Kinase 1 (S6K) is hyper-acetylated by SCFAs in T cells, leading to increased mTOR activity in T cells (24).
SCFAs can indirectly affect T cells through their effects on other cells that control T cell differentiation such as DCs (Fig. 1). SCFAs suppress the development of bone marrow progenitors into myeloid DCs in vitro (86). It has been observed that SCFAs also inhibit functional maturation of DCs in vitro (66,86,87,88,89,90). For example, C4 suppressed the maturation of bone marrow-derived DCs and production of IL-12 but increased the expression of IL-23p19 (89). Valproic acid, a branched short-chain fatty acid and potent HDAC inhibitor, suppressed the maturation of human DCs in vitro, inhibiting the up-regulation of T-cell activating molecules such as MHC II, CD80, CD86 and IL-12 (90). While the functional importance is yet to be determined, a report indicates that C4 increased CD1d at the expense of CD1a expression on developing human DCs (88). GPR109a activation affects colonic macrophages and DCs for generation of Tregs and IL-10-producing T cells (91). This effect, however, is not solely due to C4, because GPR109a is a receptor for niacin as well. In this regard, niacin treatment suppressed colitis and colon cancer in a Gpr109a-dependent manner. Moreover, Gpr109a-/- colonic epithelial cells were defective in producing IL-18 in response to C4. More studies are required to separate the niacin from SCFA effect in regulation of GPR109a. Overall, the published results indicate that the regulatory effects of SCFAs have the potential to steer DC development into tolerogenic DCs for promotion of immune tolerance. A caveat is that it remains to be fully determined if SCFAs would exert the same inhibitory effect on DCs in vivo.
The intestine is the first organ that encounters gut commensal bacteria-derived SCFAs. Therefore, SCFAs have been studied for decades for their effects on inflammatory bowel diseases (IBD). Despite some conflicting reports, high SCFA-producing conditions formed with high levels of dietary fibers are linked to decreased tissue inflammation in the intestine (92,93,94). Oral administration of C4 ameliorated T cell-induced colitis in lymphopenic mice (26). C4 administration attenuated inflammation and mucosal lesions in dextran sodium sulfate (DSS)-induced colitis, an experimental model frequently used for ulcerative colitis (95). However, there is a conflicting report that C4 administration via drinking water worsened the colitis induced by DSS (89). SCFAs also failed to regulate the acute colitis induced with 2,4,6-Trinitrobenzenesulfonic acid (TNBS) (96). These conflicting results may have been obtained due to differences in methods to induce inflammation and regimens to treat the heterogeneous inflammation. To make the function of SCFAs even more complicated, both increased and decreased DSS-induced inflammation in GPR43-deficient mice has been reported (45,97). GPR43-deficient mice had exacerbated inflammation in animal models of colitis, arthritis and asthma (97). GPR43 may modulate gut inflammation, in part, through cytokine production by mononuclear cells (98). GPR43 and GPR41, expressed by tissue cells such as epithelial cells, are also important to prevent chronic inflammation in the intestine following C. rodentium infection (40). Thus, the available information suggests that SCFA receptors play an overall beneficial role in prevention of inflammation (Fig. 2). More work is required to identify the cell types and mechanisms that mediate the beneficial effect of SCFAs in a SCFA receptor-dependent manner.
In humans, C4 enemas had a small ameliorating effect on human colitis patients (99). Treatment of patients with distal ulcerative colitis with C4 (100 mM) was effective in ameliorating disease activity (100). Moreover, treatment of patients with mild to moderate distal ulcerative colitis with combined SCFA enemas (100 mL, twice daily enemas of sodium acetate 80 mM, sodium propionate 30 mM, and sodium butyrate 40 mM) were effective in ameliorating colitis (101). A similar therapeutic effect was observed in ~50% of ulcerative colitis patients who were refractory to a rectal and oral therapy with 5-aminosalicylic acid and corticosteroid (101). SCFAs improved the efficacy of other treatments such as oral mesalazine therapy (102). There is a report that patients with mild to moderate ileocolonic Crohn's disease who were treated with 4 g/day C4 tablets for 8 weeks had decreased clinical activity (103). A caveat is that several large randomized studies found no significant effects of SCFA therapies on ulcerative colitis patients (104,105). These mixed results indicate that SCFAs and their receptors may regulate inflammatory responses only in certain pathological conditions, ameliorating certain types of inflammatory responses while exacerbating other types of responses. Beyond inflammatory bowel diseases, high fiber diets and SCFAs have suppressive effects on respiratory allergic diseases (106). Overall, SCFAs have the potential to work through multiple cell types, including T cells, to exert their regulatory effects on tissue inflammation (Fig. 2).
The gut microbial metabolites SCFAs profoundly regulate T cell differentiation in the body. Because these metabolites are produced at high levels in the gut, the T cells in the intestine and gut-associated lymphoid tissues are an important cell target for regulation by SCFAs. SCFAs can be transported into the blood and have the potential to regulate T cell activity in systemic tissue sites as well. Beyond T cells, SCFAs regulate the function and phenotype of a number of immunologically important cell types such as epithelial cells, neutrophils, and antigen presenting cells. While the anti-inflammatory activity of SCFAs has been emphasized, SCFAs can also promote the generation of effector T cells and enhance gut barrier function and innate immunity. All of these effects of SCFAs are important to maintain a healthy immune system and to prevent inflammatory diseases. More studies are required to sort out the detailed mechanism of SCFA-mediated regulation of T cells and other immune cells. The current body of literature indicates that SCFAs are not a panacea for inflammatory diseases and may exacerbate certain types of tissue inflammation. Therefore, it is important to identify the types of cells, immune responses, tissue inflammation, and diseases that are highly responsive to SCFA-based therapies.
Figures and Tables
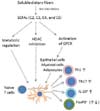 | Figure 1Regulation of T cells by SCFAs. SCFAs are actively produced by anaerobic microbiota in the colon as fermentation products of dietary materials. Most carbohydrates and proteins are completely digested and absorbed in the stomach and small intestine, and don't efficiently make SCFAs. SCFAs are mainly produced from digestion-resistant dietary fibers that reach the colon to be processed by the microbiota. SCFAs are absorbed or transported into colonocytes. They are metabolized in colonocytes or transported into blood circulation to reach other organs such as the liver and muscles. SCFAs exert their regulatory effects on epithelial cells, antigen presenting cells and T cells. Multiple mechanisms are involved including metabolic regulation, HDAC inhibition, and GPCR activation by SCFAs. These activation signals are combined to regulate T cell differentiation directly or indirectly. The direct effect of SCFAs on T cells enhances the generation of Th1 and Th17 cells in appropriate cytokine conditions, which is important to boost immunity to fight pathogens. SCFAs efficiently promote T cell production of IL-10, which is important to prevent inflammatory responses. It has been reported that SCFAs can expand FoxP3+ T cells in certain activation conditions. SCFAs may exert their regulatory effects on developing DCs to generate DCs that are limited in their ability to present antigens and cytokines to make effector T cells. These effects are combined to create the overall tolerogenic gut environment with a strong barrier function. |
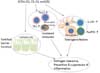 | Figure 2Regulation of tissue inflammation by SCFAs. SCFAs have the potential to regulate tissue inflammation through their effects on multiple cell types. The first cell type that is regulated by SCFAs is intestinal epithelial cells. SCFAs condition these cells to produce immune mediators that enhance the gut barrier function. Also, the response to pathogens and commensal bacteria is heightened by SCFAs during inflammatory responses. The next cell type that is affected is antigen presenting cells. SCFAs act on DCs to limit the expression of T cell-activating molecules such as MHC II molecules, co-stimulatory molecules, CCR7 and cytokines, leading to generation of tolerogenic T cells rather than inflammatory T cells. The tolerogenic effect of SCFAs on DCs can lower inflammatory responses. SCFAs can directly affect naïve T cells to steer their differentiation into both effector and IL-10-producing T cells. Moreover, SCFAs attract neutrophils to the gut during immune responses. Together, the enhanced barrier function, T cell immunity, and neutrophil recruitment help prevent infection by pathogens and invasion by commensal bacteria. By the same token, the activating activity of SCFAs for the immune cells and epithelial cells may boost inflammatory responses, if not properly regulated. |
ACKNOWLEDGEMENTS
The authors thank Kim's laboratory members for their input and assistance in preparation of this review. This study was supported, in part, from grants from NIH (R01AI074745, R01DK076616, 1R01AI080769, R21AI105620, and 1S10RR028293), USDA-NIFA, and the National Multiple Sclerosis Society to CHK.
References
1. Heinonen KM, Perreault C. Development and functional properties of thymic and extrathymic T lymphocytes. Crit Rev Immunol. 2008; 28:441–466.

2. Bhandoola A, von BH, Petrie HT, Zuniga-Pflucker JC. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity. 2007; 26:678–689.

4. Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d-restricted antigens by natural killer T cells. Nat Rev Immunol. 2012; 12:845–857.

6. Li P, Spolski R, Liao W, Leonard WJ. Complex interactions of transcription factors in mediating cytokine biology in T cells. Immunol Rev. 2014; 261:141–156.

7. Gratz IK, Campbell DJ. Organ-specific and memory treg cells: specificity, development, function, and maintenance. Front Immunol. 2014; 5:333.

8. Liston A, Gray DH. Homeostatic control of regulatory T cell diversity. Nat Rev Immunol. 2014; 14:154–165.

9. Liu X, Nurieva RI, Dong C. Transcriptional regulation of follicular T-helper (Tfh) cells. Immunol Rev. 2013; 252:139–145.

10. Tripathi SK, Lahesmaa R. Transcriptional and epigenetic regulation of T-helper lineage specification. Immunol Rev. 2014; 261:62–83.

11. Bonelli M, Shih HY, Hirahara K, Singelton K, Laurence A, Poholek A, Hand T, Mikami Y, Vahedi G, Kanno Y, O'Shea JJ. Helper T cell plasticity: impact of extrinsic and intrinsic signals on transcriptomes and epigenomes. Curr Top Microbiol Immunol. 2014; 381:279–326.

12. Kara EE, Comerford I, Fenix KA, Bastow CR, Gregor CE, McKenzie DR, McColl SR. Tailored immune responses: novel effector helper T cell subsets in protective immunity. PLoS Pathog. 2014; 10:e1003905.

13. Man K, Miasari M, Shi W, Xin A, Henstridge DC, Preston S, Pellegrini M, Belz GT, Smyth GK, Febbraio MA, Nutt SL, Kallies A. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol. 2013; 14:1155–1165.

14. Nakayama T, Yamashita M. The TCR-mediated signaling pathways that control the direction of helper T cell differentiation. Semin Immunol. 2010; 22:303–309.

15. Nurieva RI, Liu X, Dong C. Yin-Yang of costimulation: crucial controls of immune tolerance and function. Immunol Rev. 2009; 229:88–100.

16. Ishii N, Takahashi T, Soroosh P, Sugamura K. OX40-OX40 ligand interaction in T-cell-mediated immunity and immunopathology. Adv Immunol. 2010; 105:63–98.

17. Ford ML, Larsen CP. Translating costimulation blockade to the clinic: lessons learned from three pathways. Immunol Rev. 2009; 229:294–306.

18. Mace TA, King SA, Ameen Z, Elnaggar O, Young G, Riedl KM, Schwartz SJ, Clinton SK, Knobloch TJ, Weghorst CM, Lesinski GB. Bioactive compounds or metabolites from black raspberries modulate T lymphocyte proliferation, myeloid cell differentiation and Jak/STAT signaling. Cancer Immunol Immunother. 2014; 63:889–900.

19. Nicolaou A, Mauro C, Urquhart P, Marelli-Berg F. Polyunsaturated Fatty Acid-derived lipid mediators and T cell function. Front Immunol. 2014; 5:75.

20. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013; 504:451–455.

21. Benson MJ, Pino-Lagos K, Rosemblatt M, Noelle RJ. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J Exp Med. 2007; 204:1765–1774.

22. Kang SG, Lim HW, Andrisani OM, Broxmeyer HE, Kim CH. Vitamin A metabolites induce gut-homing FoxP3+ regulatory T cells. J Immunol. 2007; 179:3724–3733.

23. Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, Cheroutre H. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007; 317:256–260.

24. Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, Kim CH. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2014; doi: 10.1038/mi.2014.44.

25. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly Y, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013; 341:569–573.

26. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013; 504:446–450.

27. Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003; 62:67–72.

28. Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, Flint HJ. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000; 66:1654–1661.

29. Charrier C, Duncan GJ, Reid MD, Rucklidge GJ, Henderson D, Young P, Russell VJ, Aminov RI, Flint HJ, Louis P. A novel class of CoA-transferase involved in short-chain fatty acid metabolism in butyrate-producing human colonic bacteria. Microbiology. 2006; 152:179–185.

30. Miller TL, Wolin MJ. Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Appl Environ Microbiol. 1996; 62:1589–1592.

31. Reichardt N, Duncan SH, Young P, Belenguer A, McWilliam LC, Scott KP, Flint HJ, Louis P. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014; 8:1323–1335.

32. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014; 12:661–672.

33. Li H, Myeroff L, Smiraglia D, Romero MF, Pretlow TP, Kasturi L, Lutterbaugh J, Rerko RM, Casey G, Issa JP, Willis J, Willson JK, Plass C, Markowitz SD. SLC5A8, a sodium transporter, is a tumor suppressor gene silenced by methylation in human colon aberrant crypt foci and cancers. Proc Natl Acad Sci U S A. 2003; 100:8412–8417.

34. Miyauchi S, Gopal E, Fei YJ, Ganapathy V. Functional identification of SLC5A8, a tumor suppressor down-regulated in colon cancer, as a Na(+)-coupled transporter for short-chain fatty acids. J Biol Chem. 2004; 279:13293–13296.

35. Yanase H, Takebe K, Nio-Kobayashi J, Takahashi-Iwanaga H, Iwanaga T. Cellular expression of a sodium-dependent monocarboxylate transporter (Slc5a8) and the MCT family in the mouse kidney. Histochem Cell Biol. 2008; 130:957–966.

36. Halestrap AP, Wang X, Poole RC, Jackson VN, Price NT. Lactate transport in heart in relation to myocardial ischemia. Am J Cardiol. 1997; 80:17A–25A.

37. Eberle JA, Widmayer P, Breer H. Receptors for short-chain fatty acids in brush cells at the "gastric groove". Front Physiol. 2014; 5:152.

38. Tazoe H, Otomo Y, Karaki S, Kato I, Fukami Y, Terasaki M, Kuwahara A. Expression of short-chain fatty acid receptor GPR41 in the human colon. Biomed Res. 2009; 30:149–156.

39. Nohr MK, Pedersen MH, Gille A, Egerod KL, Engelstoft MS, Husted AS, Sichlau RM, Grunddal KV, Poulsen SS, Han S, Jones RM, Offermanns S, Schwartz TW. GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology. 2013; 154:3552–3564.

40. Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013; 145:396–406.

41. Wang A, Akers RM, Jiang H. Short communication: Presence of G protein-coupled receptor 43 in rumen epithelium but not in the islets of Langerhans in cattle. J Dairy Sci. 2012; 95:1371–1375.

42. Xiong Y, Miyamoto N, Shibata K, Valasek MA, Motoike T, Kedzierski RM, Yanagisawa M. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc Natl Acad Sci U S A. 2004; 101:1045–1050.

43. Zaibi MS, Stocker CJ, O'Dowd J, Davies A, Bellahcene M, Cawthorne MA, Brown AJ, Smith DM, Arch JR. Roles of GPR41 and GPR43 in leptin secretory responses of murine adipocytes to short chain fatty acids. FEBS Lett. 2010; 584:2381–2386.

44. Bahar HK, Veprik A, Rubins N, Naaman O, Walker MD. GPR41 gene expression is mediated by internal ribosome entry site (IRES)-dependent translation of bicistronic mRNA encoding GPR40 and GPR41 proteins. J Biol Chem. 2012; 287:20154–20163.

45. Sina C, Gavrilova O, Forster M, Till A, Derer S, Hildebrand F, Raabe B, Chalaris A, Scheller J, Rehmann A, Franke A, Ott S, Hasler R, Nikolaus S, Folsch UR, Rose-John S, Jiang HP, Li J, Schreiber S, Rosenstiel P. G protein-coupled receptor 43 is essential for neutrophil recruitment during intestinal inflammation. J Immunol. 2009; 183:7514–7522.

46. Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, Pike NB, Strum JC, Steplewski KM, Murdock PR, Holder JC, Marshall FH, Szekeres PG, Wilson S, Ignar DM, Foord SM, Wise A, Dowell SJ. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003; 278:11312–11319.

47. Voltolini C, Battersby S, Etherington SL, Petraglia F, Norman JE, Jabbour HN. A novel antiinflammatory role for the short-chain fatty acids in human labor. Endocrinology. 2012; 153:395–403.

48. Thangaraju M, Cresci GA, Liu K, Ananth S, Gnanaprakasam JP, Browning DD, Mellinger JD, Smith SB, Digby GJ, Lambert NA, Prasad PD, Ganapathy V. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009; 69:2826–2832.

49. Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, Brunet I, Wan LX, Rey F, Wang T, Firestein SJ, Yanagisawa M, Gordon JI, Eichmann A, Peti-Peterdi J, Caplan MJ. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci U S A. 2013; 110:4410–4415.

50. McCrudden FH, Fales HL. The cause of the excessive calcium excretion through the feces in infantilism. J Exp Med. 1913; 17:24–28.

51. Zoller HF, Clark WM. The production of volatile fatty acids by bacteria of the dysentery group. J Gen Physiol. 1921; 3:325–330.

52. Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001; 81:1031–1064.

53. Finnie IA, Dwarakanath AD, Taylor BA, Rhodes JM. Colonic mucin synthesis is increased by sodium butyrate. Gut. 1995; 36:93–99.

54. Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014; 121:91–119.

55. Tazoe H, Otomo Y, Kaji I, Tanaka R, Karaki SI, Kuwahara A. Roles of short-chain fatty acids receptors, GPR41 and GPR43 on colonic functions. J Physiol Pharmacol. 2008; 59:Suppl 2. 251–262.
56. Wang A, Gu Z, Heid B, Akers RM, Jiang H. Identification and characterization of the bovine G protein-coupled receptor GPR41 and GPR43 genes. J Dairy Sci. 2009; 92:2696–2705.

57. Tazoe H, Otomo Y, Karaki S, Kato I, Fukami Y, Terasaki M, Kuwahara A. Expression of short-chain fatty acid receptor GPR41 in the human colon. Biomed Res. 2009; 30:149–156.

58. Le PE, Loison C, Struyf S, Springael JY, Lannoy V, Decobecq ME, Brezillon S, Dupriez V, Vassart G, Van DJ, Parmentier M, Detheux M. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem. 2003; 278:25481–25489.

59. Vinolo MA, Ferguson GJ, Kulkarni S, Damoulakis G, Anderson K, Bohlooly Y, Stephens L, Hawkins PT, Curi R. SCFAs induce mouse neutrophil chemotaxis through the GPR43 receptor. PLoS One. 2011; 6:e21205.

60. Cani PD, Everard A, Duparc T. Gut microbiota, enteroendocrine functions and metabolism. Curr Opin Pharmacol. 2013; 13:935–940.

61. Licciardi PV, Ververis K, Karagiannis TC. Histone deacetylase inhibition and dietary short-chain Fatty acids. ISRN Allergy. 2011; 2011:869647.

62. Yin L, Laevsky G, Giardina C. Butyrate suppression of colonocyte NF-kappa B activation and cellular proteasome activity. J Biol Chem. 2001; 276:44641–44646.

63. Eftimiadi C, Buzzi E, Tonetti M, Buffa P, Buffa D, van Steenbergen MT, de GJ, Botta GA. Short-chain fatty acids produced by anaerobic bacteria alter the physiological responses of human neutrophils to chemotactic peptide. J Infect. 1987; 14:43–53.

64. Carretta MD, Conejeros I, Hidalgo MA, Burgos RA. Propionate induces the release of granules from bovine neutrophils. J Dairy Sci. 2013; 96:2507–2520.

65. Luhrs H, Gerke T, Muller JG, Melcher R, Schauber J, Boxberge F, Scheppach W, Menzel T. Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand J Gastroenterol. 2002; 37:458–466.
66. Millard AL, Mertes PM, Ittelet D, Villard F, Jeannesson P, Bernard J. Butyrate affects differentiation, maturation and function of human monocyte-derived dendritic cells and macrophages. Clin Exp Immunol. 2002; 130:245–255.

67. Park JS, Lee EJ, Lee JC, Kim WK, Kim HS. Anti-inflammatory effects of short chain fatty acids in IFN-gamma-stimulated RAW 264.7 murine macrophage cells: involvement of NF-kappaB and ERK signaling pathways. Int Immunopharmacol. 2007; 7:70–77.

68. Kendrick SF, O'Boyle G, Mann J, Zeybel M, Palmer J, Jones DE, Day CP. Acetate, the key modulator of inflammatory responses in acute alcoholic hepatitis. Hepatology. 2010; 51:1988–1997.

69. Arora T, Sharma R, Frost G. Propionate. Anti-obesity and satiety enhancing factor? Appetite. 2011; 56:511–515.

70. Hong YH, Nishimura Y, Hishikawa D, Tsuzuki H, Miyahara H, Gotoh C, Choi KC, Feng DD, Chen C, Lee HG, Katoh K, Roh SG, Sasaki S. Acetate and propionate short chain fatty acids stimulate adipogenesis via GPCR43. Endocrinology. 2005; 146:5092–5099.

71. Ge H, Li X, Weiszmann J, Wang P, Baribault H, Chen JL, Tian H, Li Y. Activation of G protein-coupled receptor 43 in adipocytes leads to inhibition of lipolysis and suppression of plasma free fatty acids. Endocrinology. 2008; 149:4519–4526.

72. Kimura I, Inoue D, Maeda T, Hara T, Ichimura A, Miyauchi S, Kobayashi M, Hirasawa A, Tsujimoto G. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41). Proc Natl Acad Sci U S A. 2011; 108:8030–8035.

73. Nancey S, Bienvenu J, Coffin B, Andre F, Descos L, Flourie B. Butyrate strongly inhibits in vitro stimulated release of cytokines in blood. Dig Dis Sci. 2002; 47:921–928.
74. Cavaglieri CR, Nishiyama A, Fernandes LC, Curi R, Miles EA, Calder PC. Differential effects of short-chain fatty acids on proliferation and production of pro- and anti-inflammatory cytokines by cultured lymphocytes. Life Sci. 2003; 73:1683–1690.

75. Kurita-Ochiai T, Fukushima K, Ochiai K. Volatile fatty acids, metabolic by-products of periodontopathic bacteria, inhibit lymphocyte proliferation and cytokine production. J Dent Res. 1995; 74:1367–1373.

76. Zimmerman MA, Singh N, Martin PM, Thangaraju M, Ganapathy V, Waller JL, Shi H, Robertson KD, Munn DH, Liu K. Butyrate suppresses colonic inflammation through HDAC1-dependent Fas upregulation and Fas-mediated apoptosis of T cells. Am J Physiol Gastrointest Liver Physiol. 2012; 302:G1405–G1415.

77. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013; 504:446–450.

78. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013; 500:232–236.

79. Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G. Mammalian TOR: a homeostatic ATP sensor. Science. 2001; 294:1102–1105.

80. Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009; 30:832–844.

81. Chen S, Liu D, Wu J, Xu B, Lu K, Zhu W, Chen M. Effect of inhibiting the signal of mammalian target of rapamycin on memory T cells. Transplant Proc. 2014; 46:1642–1648.

82. Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. 2002; 132:1012–1017.

83. Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009; 10:32–42.

84. Yu X, Shahir AM, Sha J, Feng Z, Eapen B, Nithianantham S, Das B, Karn J, Weinberg A, Bissada NF, Ye F. Short-chain fatty acids from periodontal pathogens suppress histone deacetylases, EZH2, and SUV39H1 to promote Kaposi's sarcoma-associated herpesvirus replication. J Virol. 2014; 88:4466–4479.

85. Fenton TR, Gwalter J, Ericsson J, Gout IT. Histone acetyltransferases interact with and acetylate p70 ribosomal S6 kinases in vitro and in vivo. Int J Biochem Cell Biol. 2010; 42:359–366.

86. Singh N, Thangaraju M, Prasad PD, Martin PM, Lambert NA, Boettger T, Offermanns S, Ganapathy V. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J Biol Chem. 2010; 285:27601–27608.

87. Wang B, Morinobu A, Horiuchi M, Liu J, Kumagai S. Butyrate inhibits functional differentiation of human monocyte-derived dendritic cells. Cell Immunol. 2008; 253:54–58.

88. Nascimento CR, Freire-de-Lima CG, da Silva de OA, Rumjanek FD, Rumjanek VM. The short chain fatty acid sodium butyrate regulates the induction of CD1a in developing dendritic cells. Immunobiology. 2011; 216:275–284.

89. Berndt BE, Zhang M, Owyang SY, Cole TS, Wang TW, Luther J, Veniaminova NA, Merchant JL, Chen CC, Huffnagle GB, Kao JY. Butyrate increases IL-23 production by stimulated dendritic cells. Am J Physiol Gastrointest Liver Physiol. 2012; 303:G1384–G1392.

90. Frikeche J, Simon T, Brissot E, Gregoire M, Gaugler B, Mohty M. Impact of valproic acid on dendritic cells function. Immunobiology. 2012; 217:704–710.

91. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, Lee JR, Offermanns S, Ganapathy V. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014; 40:128–139.

92. Ananthakrishnan AN, Khalili H, Konijeti GG, Higuchi LM, de SP, Korzenik JR, Fuchs CS, Willett WC, Richter JM, Chan AT. A prospective study of long-term intake of dietary fiber and risk of Crohn's disease and ulcerative colitis. Gastroenterology. 2013; 145:970–977.

93. Amre DK, D'Souza S, Morgan K, Seidman G, Lambrette P, Grimard G, Israel D, Mack D, Ghadirian P, Deslandres C, Chotard V, Budai B, Law L, Levy E, Seidman EG. Imbalances in dietary consumption of fatty acids, vegetables, and fruits are associated with risk for Crohn's disease in children. Am J Gastroenterol. 2007; 102:2016–2025.

94. Hou JK, Abraham B, El-Serag H. Dietary intake and risk of developing inflammatory bowel disease: systematic review of the literature. Am J Gastroenterol. 2011; 106:563–573.

95. Vieira EL, Leonel AJ, Sad AP, Beltrao NR, Costa TF, Ferreira TM, Gomes-Santos AC, Faria AM, Peluzio MC, Cara DC, varez-Leite JI. Oral administration of sodium butyrate attenuates inflammation and mucosal lesion in experimental acute ulcerative colitis. J Nutr Biochem. 2012; 23:430–436.

96. Tarrerias AL, Millecamps M, Alloui A, Beaughard C, Kemeny L, Bourdu S, Bommelaer G, Eschalier A, Dapoigny M, Ardid D. Short-chain fatty acid enemas fail to decrease colonic hypersensitivity and inflammation in TNBS-induced colonic inflammation in rats. Pain. 2002; 100:91–97.

97. Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009; 461:1282–1286.

98. Masui R, Sasaki M, Funaki Y, Ogasawara N, Mizuno M, Iida A, Izawa S, Kondo Y, Ito Y, Tamura Y, Yanamoto K, Noda H, Tanabe A, Okaniwa N, Yamaguchi Y, Iwamoto T, Kasugai K. G protein-coupled receptor 43 moderates gut inflammation through cytokine regulation from mononuclear cells. Inflamm Bowel Dis. 2013; 19:2848–2856.

99. Hamer HM, Jonkers DM, Vanhoutvin SA, Troost FJ, Rijkers G, de BA, Bast A, Venema K, Brummer RJ. Effect of butyrate enemas on inflammation and antioxidant status in the colonic mucosa of patients with ulcerative colitis in remission. Clin Nutr. 2010; 29:738–744.

100. Scheppach W, Sommer H, Kirchner T, Paganelli GM, Bartram P, Christl S, Richter F, Dusel G, Kasper H. Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology. 1992; 103:51–56.

101. Vernia P, Marcheggiano A, Caprilli R, Frieri G, Corrao G, Valpiani D, Di Paolo MC, Paoluzi P, Torsoli A. Short-chain fatty acid topical treatment in distal ulcerative colitis. Aliment Pharmacol Ther. 1995; 9:309–313.

102. Vernia P, Monteleone G, Grandinetti G, Villotti G, Di GE, Frieri G, Marcheggiano A, Pallone F, Caprilli R, Torsoli A. Combined oral sodium butyrate and mesalazine treatment compared to oral mesalazine alone in ulcerative colitis: randomized, double-blind, placebo-controlled pilot study. Dig Dis Sci. 2000; 45:976–981.
103. Di SA, Morera R, Ciccocioppo R, Cazzola P, Gotti S, Tinozzi FP, Tinozzi S, Corazza GR. Oral butyrate for mildly to moderately active Crohn's disease. Aliment Pharmacol Ther. 2005; 22:789–794.

104. Steinhart AH, Hiruki T, Brzezinski A, Baker JP. Treatment of left-sided ulcerative colitis with butyrate enemas: a controlled trial. Aliment Pharmacol Ther. 1996; 10:729–736.

105. Breuer RI, Soergel KH, Lashner BA, Christ ML, Hanauer SB, Vanagunas A, Harig JM, Keshavarzian A, Robinson M, Sellin JH, Weinberg D, Vidican DE, Flemal KL, Rademaker AW. Short chain fatty acid rectal irrigation for left-sided ulcerative colitis: a randomised, placebo controlled trial. Gut. 1997; 40:485–491.

106. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, Marsland BJ. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014; 20:159–166.

107. Hadjiagapiou C, Schmidt L, Dudeja PK, Layden TJ, Ramaswamy K. Mechanism(s) of butyrate transport in Caco-2 cells: role of monocarboxylate transporter 1. Am J Physiol Gastrointest Liver Physiol. 2000; 279:G775–G780.

108. Alrefai WA, Tyagi S, Gill R, Saksena S, Hadjiagapiou C, Mansour F, Ramaswamy K, Dudeja PK. Regulation of butyrate uptake in Caco-2 cells by phorbol 12-myristate 13-acetate. Am J Physiol Gastrointest Liver Physiol. 2004; 286:G197–G203.

109. Ritzhaupt A, Ellis A, Hosie KB, Shirazi-Beechey SP. The characterization of butyrate transport across pig and human colonic luminal membrane. J Physiol. 1998; 507(Pt 3):819–830.

110. Gopal E, Fei YJ, Miyauchi S, Zhuang L, Prasad PD, Ganapathy V. Sodium-coupled and electrogenic transport of B-complex vitamin nicotinic acid by slc5a8, a member of the Na/glucose co-transporter gene family. Biochem J. 2005; 388:309–316.

111. Miyauchi S, Gopal E, Babu E, Srinivas SR, Kubo Y, Umapathy NS, Thakkar SV, Ganapathy V, Prasad PD. Sodium-coupled electrogenic transport of pyroglutamate (5-oxoproline) via SLC5A8, a monocarboxylate transporter. Biochim Biophys Acta. 2010; 1798:1164–1171.

112. Thangaraju M, Cresci G, Itagaki S, Mellinger J, Browning DD, Berger FG, Prasad PD, Ganapathy V. Sodium-coupled transport of the short chain fatty acid butyrate by SLC5A8 and its relevance to colon cancer. J Gastrointest Surg. 2008; 12:1773–1781.

113. Gopal E, Fei YJ, Sugawara M, Miyauchi S, Zhuang L, Martin P, Smith SB, Prasad PD, Ganapathy V. Expression of slc5a8 in kidney and its role in Na(+)-coupled transport of lactate. J Biol Chem. 2004; 279:44522–44532.

114. Martin PM, Dun Y, Mysona B, Ananth S, Roon P, Smith SB, Ganapathy V. Expression of the sodium-coupled monocarboxylate transporters SMCT1 (SLC5A8) and SMCT2 (SLC5A12) in retina. Invest Ophthalmol Vis Sci. 2007; 48:3356–3363.

115. Martin PM, Gopal E, Ananth S, Zhuang L, Itagaki S, Prasad BM, Smith SB, Prasad PD, Ganapathy V. Identity of SMCT1 (SLC5A8) as a neuron-specific Na+-coupled transporter for active uptake of L-lactate and ketone bodies in the brain. J Neurochem. 2006; 98:279–288.

116. Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, Cameron J, Grosse J, Reimann F, Gribble FM. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012; 61:364–371.

117. Hong YH, Nishimura Y, Hishikawa D, Tsuzuki H, Miyahara H, Gotoh C, Choi KC, Feng DD, Chen C, Lee HG, Katoh K, Roh SG, Sasaki S. Acetate and propionate short chain fatty acids stimulate adipogenesis via GPCR43. Endocrinology. 2005; 146:5092–5099.

118. Dewulf EM, Ge Q, Bindels LB, Sohet FM, Cani PD, Brichard SM, Delzenne NM. Evaluation of the relationship between GPR43 and adiposity in human. Nutr Metab (Lond). 2013; 10:11.

119. Tang Y, Chen Y, Jiang H, Robbins GT, Nie D. G-protein-coupled receptor for short-chain fatty acids suppresses colon cancer. Int J Cancer. 2011; 128:847–856.

120. Karaki S, Mitsui R, Hayashi H, Kato I, Sugiya H, Iwanaga T, Furness JB, Kuwahara A. Short-chain fatty acid receptor, GPR43, is expressed by enteroendocrine cells and mucosal mast cells in rat intestine. Cell Tissue Res. 2006; 324:353–360.

121. Nilsson NE, Kotarsky K, Owman C, Olde B. Identification of a free fatty acid receptor, FFA2R, expressed on leukocytes and activated by short-chain fatty acids. Biochem Biophys Res Commun. 2003; 303:1047–1052.

122. Wanders D, Graff EC, Judd RL. Effects of high fat diet on GPR109A and GPR81 gene expression. Biochem Biophys Res Commun. 2012; 425:278–283.

123. Taggart AK, Kero J, Gan X, Cai TQ, Cheng K, Ippolito M, Ren N, Kaplan R, Wu K, Wu TJ, Jin L, Liaw C, Chen R, Richman J, Connolly D, Offermanns S, Wright SD, Waters MG. (D)-beta-Hydroxybutyrate inhibits adipocyte lipolysis via the nicotinic acid receptor PUMA-G. J Biol Chem. 2005; 280:26649–26652.

124. Ingersoll MA, Potteaux S, Alvarez D, Hutchison SB, van RN, Randolph GJ. Niacin inhibits skin dendritic cell mobilization in a GPR109A independent manner but has no impact on monocyte trafficking in atherosclerosis. Immunobiology. 2012; 217:548–557.

125. Li X, Millar JS, Brownell N, Briand F, Rader DJ. Modulation of HDL metabolism by the niacin receptor GPR109A in mouse hepatocytes. Biochem Pharmacol. 2010; 80:1450–1457.

126. Bermudez Y, Benavente CA, Meyer RG, Coyle WR, Jacobson MK, Jacobson EL. Nicotinic acid receptor abnormalities in human skin cancer: implications for a role in epidermal differentiation. PLoS One. 2011; 6:e20487.

127. Xu LL, Stackhouse BG, Florence K, Zhang W, Shanmugam N, Sesterhenn IA, Zou Z, Srikantan V, Augustus M, Roschke V, Carter K, McLeod DG, Moul JW, Soppett D, Srivastava S. PSGR, a novel prostate-specific gene with homology to a G protein-coupled receptor, is overexpressed in prostate cancer. Cancer Res. 2000; 60:6568–6572.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download