Abstract
T cell immunoglobulin mucin domain (TIM)-3 is an immunomodulatory molecule and upregulated in T cells by several cytokines. TIM-3 also influences mast cell function but its transcriptional regulation in mast cells has not been clarified. Therefore, we examined the transcript level and the promoter activity of TIM-3 in mast cells. The TIM-3 transcript level was assessed by real-time RT-PCR and promoter activity by luciferase reporter assay. TIM-3 mRNA levels were increased in HMC-1, a human mast cell line by TGF-β1 stimulation but not by stimulation with interferon (IFN)-α, IFN-λ, TNF-α, or IL-10. TIM-3 promoter -349~+144 bp region relative to the transcription start site was crucial for the basal and TGF-β1-induced TIM-3 promoter activities in HMC-1 cells. TIM-3 promoter activity was increased by overexpression of Smad2 and Smad4, downstream molecules of TGF-β1 signaling. Our results localize TIM-3 promoter activity to the region spanning -349 to +144 bp in resting and TGF-β1 stimulated mast cells.
T cell immunoglobulin mucin domain (TIM)-3 is expressed in various leukocyte subpopulations and modulates their functions (1). It is expressed on the surfaces of exhausted T cells and involved in the downregulation of effector function of T cells (2-4). TIM-3 on dendritic cells promotes the uptake of apoptotic cells via interaction with phosphatidylserine but suppresses the immunogenicity of nucleic acid via interaction with HMGB1 (5,6). TIM-3 expression in mast cells influences cytokine production and apoptosis of these cells (7). However, the regulation of TIM-3 expression has not been well known.
Mast cells play a significant role in various immune responses. These cells lead allergic symptoms through secretion of mediators and cytokines upon cross-linking of FcεRI (8). Mast cells contribute defense against pathogen; mice devoid of mast cells die from infection that does not result in death in wild type mice (9). Protective immunity is enhanced by mast cell production of TNF-α and recruitment of neutrophils to the infectious site (9). Mast cells are also required for the development of collagen-induced rheumatoid arthritis and the induction of tolerance to skin graft (10,11).
To understand the regulation of TIM-3 expression in mast cells, we investigated the effect of various cytokines on TIM-3 transcription and the activity of TIM-3 promoter in relation with TGF-β1 stimulation of mast cells.
IFN-α, IFN-λ, TGF-β1, IL-10, and TNF-α were purchased from R&D Systems (Minneapolis, MN, USA). Expression vectors for Smad2 and Smad4 were kindly provided by Dr. Cho (Dept. Biochemistry, Ajou University School of Medicine, Korea).
HMC-1, a human mast cell line, was kindly provided by Professor Hyung Min, Kim (Kyunghee University, Suwon, Korea). HMC-1 cells were maintained in IMDM supplemented with 10% FBS, penicillin-streptomycin (each 100 U/ml, 100 ug/ml). Similarly, HEK293 cells were maintained in RPMI 1640 (Gibco BRL, Paisley, Scotland) supplemented with 10% FBS.
The total RNA was isolated using RNA STAT-60 (Tel-Test, IN C., Friendwood, TX, USA) and reverse transcribed using Superscriptase II (Invitrogen, Carsbade, CA, USA). Real-time PCR was performed using primers (5'-TCCAAGGATGCTTACCACC AG-3': 5'-GCCAATGTGGATATTTGTGTTAGATT-3') and a Taq Man probe (5'-ACATGGCCCAGCAGAGACACAGACACT-3') for TIM-3 transcript which was normalized to GAPDH transcript levels.
Luciferase reporter vectors were constructed by ligation of human TIM-3 promoter region DNA fragment into pGL-Basic vector (Promega, Madison, WI, USA). Human TIM-3 promoter DNA was amplified by PCR using genomic DNA isolated from HMC-1 cells. For amplification of TIM-3 -1677~+144 DNA fragment, primers TIM3+1 (5'-GGAGCTTGCAGAAGAAAAGTCAGAGGACACCTCTGTTAGG-3') and 5'-AGAGCCTTGACCAAGTTCATGCTGCTAATAAAAATAACCCCAG-3' were used. For TIM-3 -872~+144 DNA fragment, primers TIM3+1 and 5'-CTTTTGCTTTTAAGGTGTCCAGATAAAGGTCACACTCCCAG-3' were used. For TIM-3 -349~+144 DNA fragment, primers TIM3+1 and 5'-CTGTGACCAAAGTTTATGAAGCC-3' were used. The PCR products was cloned into Topo TA cloning vector (Invitorgen) and the nucleotide sequences were verified by comparison with the gene sequence (NW_001838954). Then TIM-3 promoter DNA was subcloned into pGL-Basic vector using Nhe I and Bgl II and designated as T3U(1.8)-luc, T3U(1.0)-luc and T3U(0.5)-luc.
HMC-1 cells (1×106) were transfected with 3.6µg of luciferase reporter vector together with 400 ng of pEGFP-N1 plasmid (Clonetech, Mountain View, CA, USA) using electroporator (Digital Bio Technology, Seoul, Korea). The cells were incubated for 48 h in 5% CO2 incubator at the 37℃, and then luciferase activity was analyzed using luciferin (Promega, Madison, WI, USA) and luminometer (Molecular devices, Sunnyvale, CA, USA). The transfection efficiency was analyzed by GFP-expressing cell frequencies using flow cytometer (FacsCanto). Similarly, luciferase activity was assessed in HEK293 cells transfected with DNA using Lipofectamin 2000 (Invitrogen, Carsbade, CA, USA).
In recent studies, TIM-3 mRNA and protein expression levels were up regulated in TGF-β1 stimulated-human mast cells (12) but the transcriptional regulation of TIM-3 by other stimulants has not been well explored. To address this issue a human mast cell line, HMC-1 cells were treated with various cytokines for the indicated time and then TIM-3 mRNA levels were determined by real-time RT-PCR. TIM-3 mRNA expression was not significantly enhanced by treatment with the indicated concentration of IFN-α IFN-λ, IL-10, or TNF-α (p>0.05) but was significantly increased by TGF-β1 stimulation of HMC-1 cells for 4 h (p<0.005) (Fig. 1).
To know the TIM-3 promoter is responsive to TGF-β1 stimulation, we first examined the basal activity of TIM-3 promoter by luciferase reporter assay using vectors depicted in Fig. 2A. Luciferase activities in both HEK293 cells and HMC-1 cells transfected with T3U(0.5)-luc that contains proximal TIM-3 promoter were significantly higher than that of controls transfected with the empty vector pGL-Basic (Fig. 2B and C). Also, luciferase activities driven by T3U(1.0)-luc and T3U(1.8)-luc, respectively were significantly higher than that of controls but lower than that driven by T3U(0.5)-luc. These results indicate that the proximal TIM-3 promoter spanning from +144 to -349 may consist of sufficient element for the basal level transcription of TIM-3 in HMC-1 cells.
We next analyzed the TIM-3 promoter activity in the presence of TGF-β1 stimulation (Fig. 3). Luciferase activity driven by each luciferase reporter vector in the presence of TGF-β1 was presented as fold induction relative to that in the absence of TGF-β1. Luciferase activity in HMC-1 cells transfected with T3U(0.5)-luc or T3U(1.8)-luc was significantly increased (2.3 and 2.6 fold, respectively) by TGF-β1 stimulation compared to that in the absence of TGF-β1 stimulation (p<0.001). However, luciferase activity in HMC-1 cells transfected with T3U(1.0)-luc or the empty pGL-Basic was not significantly increased by TGF-β1 stimulation compared to that in the absence of TGF-β1 stimulation (p>0.01). These results suggest that TIM-3 promoter region may contain the TGF-β1 responsive elements.
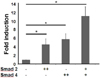
To support the TGF-β1 responsiveness of the TIM-3 promoter, we assessed TIM-3 promoter activity in the cells overexpressing Smad2 and Smad4 that were reported to be involved in the gene expression induced by TGF-β1 (13,14). Given that TGF-β responsive promoter activity was increased by overexpression of either Smad2 or Smad3 in HEK293 cells even in the absence of TGF-β stimulation (15), we analyzed TIM-3 promoter activity in HEK293 cells overexpressing Smad2 and/or Smad4 without TGF-β1 treatment (Fig. 4). Compared to control, luciferase activity was significantly increased in HEK293 cells by overexpression of Smad2, Smad4 or both (4.5, 5.8 and 11 fold, respectively) (p<0.01). These results imply that TIM-3 promoter region may respond to TGF-β1 stimulation through Smad2 and Smad4 involvement.
In this study, we revealed that TIM-3 mRNA expression in a human mast cell line was increased by TGF-β1 stimulation but not by other stimuli such as interferon α and λ, TNF-α, and IL-10. TGF-β1 affects mast cell survival and functions. TGF-β1 inhibits IL-3-dependent mast cell proliferation and counterbalances the effect of IL-4 on mast cell survival, migration, and FcεRI expression (16,17). Also TGF-β1 can elicit mouse mast cell protease-1 expression (18) and mast cell tryptase expression in experimental emphysema model (19). Furthermore, TGF-β1 in both soluble and regulatory T cell-surface bound forms can escalate IL-6 production by mast cells (20). Our results (21) and the previous report by Wiener et al.(12) add another one to the effects of TGF-β1 on mast cells i.e. induction of TIM-3 expression. Except the report by Nakae et al.(7) that TIM-3 cross-linking by an anti-TIM-3 polyclonal Ab can promote IL-4, IL-6 and IL-13 production but suppress mast cell apoptosis, little information is currently available regarding the role of TIM-3 in mast cell function.
We also demonstrated that TIM-3 promoter -349~+144 bp region relative to the transcription start site was crucial for the basal and TGF-β1-induced TIM-3 transcription in HMC-1 cells. Since T3U(0.5)-luc driven luciferase activity in HMC-1 cells was greater than that driven by T3U(1.8)-luc or T3U(1.0)-luc, the DNA fragment of -349~+144 bp seems to contain elements for basal transcription of TIM-3. Compatible to our results, Zhang et al. (22) reported that the basal TIM-3 promoter activity is localized to the region spanning -241 to + 63 bp in YT cells, a T/NK cell line. Apparently incompatible with our previous observation that TIM-3 promoter activity driven by the -1,362 to +144 bp region was not enhanced by TGF-β1 stimulation (21), luciferase activities under TIM-3 promoter -349~+144 bp region and -1,677~+144 was significantly increased in HMC-1 cells by TGF-β1 stimulation (Fig. 3). Interestingly, the luciferase activity under TIM-3 promoter -872~+144 was not significantly elevated by TGF-β 1 stimulation in the present study. Further study is required to investigate whether TIM-3 promoter -1,362 to -350 bp region may contain the element hindering the TGF-β1 responsiveness of TIM-3 promoter -349~+144 bp region.
We showed that overexpression of Smad2 and Smad4 upregulated TIM-3 promoter activity in HEK293 cells. Smad2 and Smad4 are downstream signaling molecules and transcription factors of TGF-β1 signaling (13,14). Although there are three predicted Smad binding elements in TIM-3 promoter -349~+144 bp region, further study should be followed to clarify whether Smad2 and Smad4 are directly bound to TIM-3 promoter or indirectly involved in TIM-3 transcription in TGF-β1 stimulated mast cells. In T cells, it was reported that two transcription factors regulated TIM-3 transcription in different modes. T-box transcription factor T-bet increases Tim-3 transcription via its interaction with Tim-3 promoter of mouse TH1 cells at approximately 400 bp upstream of the first ATG, whereas signal transducer and activator of transcription (STAT)-4 dose not bind to the Tim-3 promoter but reduces Tim-3 expression in murine T cells when it is knocked-out (23). At present, critical role of MEK, another downstream signaling molecule of TGF-β1 signal pathway, in TIM-3 induction is revealed in HMC-1 cells stimulated with TGF-β1 (21).
Conclusively, our results localize TIM-3 promoter activity in resting and TGF-β1 stimulated HMC-1 cells to the region from -349 to +144 bp relative to the transcription start site and propose a possible regulatory role of Smad2 and Smad4 in TIM-3 transcription of mast cells.
Figures and Tables
Figure 1
TIM-3 mRNA expression was increased in HMC-1 cells by stimulation with TGF-β1 but not by stimulation with IFNs, IL-10, and TNF-α. HMC-1 cells were treated for the indicated time with IFN-α (100 U/ml), IFN-λ (100 ng/ml), TGF-β1 (2 ng/ml), IL-10 (10 ng/ml), and TNF-α (10 ng/ml), respectively and then TIM-3 transcript levels were analyzed by real-time RT-PCR. TIM-3 transcript levels were normalized to GAPDH transcript levels. Relative Tim-3 mRNA levels to control (Con, without cytokine treatment) were presented. *p<0.005.
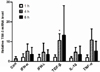
Figure 2
Basal TIM-3 promoter activity in HEK293 and HMC-1 cells. (A) Luciferase reporter vectors containing various length of TIM-3 promoter region. (B) HEK293 (5×105) or (C) HMC-1 (1×106) cells were transfected with the indicated luciferase reporter vector together with pEGFP-N1 vector (1/10 of total plasmid) and 48 h later luciferase activity was measured and normalized to the frequencies of GFP-expressing cells. Relative luciferase activity to control (pGL-Basic) was presented. The data are the mean±standard deviation of 5 experiments. *p<0.01.

Figure 3
TIM-3 promoter activity in HMC-1 cells stimulated with TGF-β1. HMC-1 (1×106) cells were transfected with the indicated luciferase reporter vector together with pEGFP-N1 vector (1/10 of total plasmid) and 42 h later treated with TGF-β1 (2 ng/ml) for 6 h. Then luciferase activity was measured and normalized to the frequencies of GFP-expressing cells. Fold induction of luciferase activity by TGF-β1 treatment was presented for each luciferase reporter vector. The data are the mean±standard deviation of 5 experiments. *p<0.001.

Figure 4
TIM-3 promoter activity was increased by Smad overexpression in HEK293 cells. HEK293 cells (1×106) were transfected with T3U(1.8)-luc (1.8µg) together with the indicated Smad expression vector (+: 0.9µg, ++: 1.8µg) or control empty vector pcDNA (1.8µg). For normalization of transfection efficiency, pEGFP-N1 vector (1/10 of total plasmid) was also co-transfected. After 48 h, the luciferase activity was measured and normalized to the frequencies of GFP-expressing cells. Fold induction of luciferase activity compared to that in the absence of Smad overexpression was presented. The data are the mean±standard deviation of 5 experiments. *p<0.01.
ACKNOWLEDGEMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (2010-0024113).
References
1. Sakuishi K, Jayaraman P, Behar SM, Anderson AC, Kuchroo VK. Emerging Tim-3 functions in antimicrobial and tumor immunity. Trends Immunol. 2011. 32:345–349.

2. Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med. 2010. 207:2187–2194.

3. Zhou Q, Munger ME, Veenstra RG, Weigel BJ, Hirashima M, Munn DH, Murphy WJ, Azuma M, Anderson AC, Kuchroo VK, Blazar BR. Coexpression of Tim-3 and PD-1 identifies a CD8+ T-cell exhaustion phenotype in mice with disseminated acute myelogenous leukemia. Blood. 2011. 117:4501–4510.

4. Lee MJ, Woo MY, Chwae YJ, Kwon MH, Kim K, Park S. Down-regulation of interleukin-2 production by CD4(+) T cells expressing TIM-3 through suppression of NFAT dephosphorylation and AP-1 transcription. Immunobiology. 2012. 217:986–995.

5. Chiba S, Baghdadi M, Akiba H, Yoshiyama H, Kinoshita I, Dosaka-Akita H, Fujioka Y, Ohba Y, Gorman JV, Colgan JD, Hirashima M, Uede T, Takaoka A, Yagita H, Jinushi M. Tumor-infiltrating DCs suppress nucleic acid-mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1. Nat Immunol. 2012. 13:832–842.

6. Nakayama M, Akiba H, Takeda K, Kojima Y, Hashiguchi M, Azuma M, Yagita H, Okumura K. Tim-3 mediates phagocytosis of apoptotic cells and cross-presentation. Blood. 2009. 113:3821–3830.

7. Nakae S, Iikura M, Suto H, Akiba H, Umetsu DT, Dekruyff RH, Saito H, Galli SJ. TIM-1 and TIM-3 enhancement of Th2 cytokine production by mast cells. Blood. 2007. 110:2565–2568.

8. Bischoff SC. Role of mast cells in allergic and non-allergic immune responses: comparison of human and murine data. Nat Rev Immunol. 2007. 7:93–104.

9. Malaviya R, Ikeda T, Ross E, Abraham SN. Mast cell modulation of neutrophil influx and bacterial clearance at sites of infection through TNF-alpha. Nature. 1996. 381:77–80.

10. Lee DM, Friend DS, Gurish MF, Benoist C, Mathis D, Brenner MB. Mast cells: a cellular link between autoantibodies and inflammatory arthritis. Science. 2002. 297:1689–1692.

11. Lu LF, Lind EF, Gondek DC, Bennett KA, Gleeson MW, Pino-Lagos K, Scott ZA, Coyle AJ, Reed JL, Van Snick J, Strom TB, Zheng XX, Noelle RJ. Mast cells are essential intermediaries in regulatory T-cell tolerance. Nature. 2006. 442:997–1002.

12. Wiener Z, Kohalmi B, Pocza P, Jeager J, Tolgyesi G, Toth S, Gorbe E, Papp Z, Falus A. TIM-3 is expressed in melanoma cells and is upregulated in TGF-beta stimulated mast cells. J Invest Dermatol. 2007. 127:906–914.

13. Schmierer B, Hill CS. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol. 2007. 8:970–982.

14. Conidi A, Cazzola S, Beets K, Coddens K, Collart C, Cornelis F, Cox L, Joke D, Dobreva MP, Dries R, Esguerra C, Francis A, Ibrahimi A, Kroes R, Lesage F, Maas E, Moya I, Pereira PN, Stappers E, Stryjewska A, van den Berghe V, Vermeire L, Verstappen G, Seuntjens E, Umans L, Zwijsen A, Huylebroeck D. Few Smad proteins and many Smad-interacting proteins yield multiple functions and action modes in TGFβ/BMP signaling in vivo. Cytokine Growth Factor Rev. 2011. 22:287–300.

15. Soond SM, Chantry A. Selective targeting of activating and inhibitory Smads by distinct WWP2 ubiquitin ligase isoforms differentially modulates TGFβ signalling and EMT. Oncogene. 2011. 30:2451–2462.

16. Broide DH, Wasserman SI, Alvaro-Gracia J, Zvaifler NJ, Firestein GS. Transforming growth factor-beta 1 selectively inhibits IL-3-dependent mast cell proliferation without affecting mast cell function or differentiation. J Immunol. 1989. 143:1591–1597.
17. Macey MR, Sturgill JL, Morales JK, Falanga YT, Morales J, Norton SK, Yerram N, Shim H, Fernando J, Gifillan AM, Gomez G, Schwartz L, Oskeritzian C, Spiegel S, Conrad D, Ryan JJ. IL-4 and TGF-beta 1 counterbalance one another while regulating mast cell homeostasis. J Immunol. 2010. 184:4688–4695.

18. Miller HR, Wright SH, Knight PA, Thornton EM. A novel function for transforming growth factor-beta1: upregulation of the expression and the IgE-independent extracellular release of a mucosal mast cell granule-specific beta-chymase, mouse mast cell protease-1. Blood. 1999. 93:3473–3486.

19. Mortaz E, Givi ME, Da Silva CA, Folkerts G, Redegeld FA. A relation between TGF-β and mast cell tryptase in experimental emphysema models. Biochim Biophys Acta. 2012. 1822:1154–1160.

20. Ganeshan K, Bryce PJ. Regulatory T cells enhance mast cell production of IL-6 via surface-bound TGF-β. J Immunol. 2012. 188:594–603.

21. Yoon SJ, Lee MJ, Shin DC, Kim JS, Chwae YJ, Kwon MH, Kim K, Park S. Activation of mitogen activated protein kinase-Erk kinase (MEK) increases T cell immunoglobulin mucin domain-3 (TIM-3) transcription in human T lymphocytes and a human mast cell line. Mol Immunol. 2011. 48:1778–1783.

22. Zhang J, Daley D, Akhabir L, Stefanowicz D, Chan-Yeung M, Becker AB, Laprise C, Paré PD, Sandford AJ. Lack of association of TIM3 polymorphisms and allergic phenotypes. BMC Med Genet. 2009. 10:62.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download