Abstract
Background
Epstein-Barr virus associated gastric lymphoepithelioma-like carcinoma (LELC) is characterized by the intensive infiltration of lymphoid cells, the presence of EBV, and the better prognosis over typical adenocarcinoma. Thus, it was assumable that viral latent proteins may be responsible for the recruitment of a certain T cell repertoire to EBV-associated gastric carcinoma.
Methods
To examine above possibility, EBV gene expression in gastric carcinoma tissues and usage of TCR among the tumor infiltrating lymphocytes were analyzed.
Results
EBV specific DNA and EBERs RNA were detected in 4 out of 30 patients. RT-PCR analysis revealed that all 4 of EBV-positive tumor tissues expressed EBNA1 mRNA and BARTs and LMP2a was detected only one sample out of 4. However, the EBNA2 and LMP-1 transcripts were not detected in these tissues. CD8+ T cells were the predominant population of infiltrating lymphocytes in the EBV-positive gastric carcinoma. According to spectra type analysis of infiltrating T cells, 10 predominant bands were detected by TCR Vβ CDR3 specific RT-PCR from 4 EBV-positive tumor tissues. Sequence analysis of these bands revealed oligoclonal expansion of T cells.
Epstein-Barr virus (EBV) is a DNA virus that belongs to the Herpesviridae family and infects more than 90% of the entire world population. EBV causes infectious mononucleosis, and is related to lymphocyte and epithelial cell origin tumors such as Burkitt's lymphoma, lymphomas associated with immunocompromised patients, and undifferentiated nasopharyngeal carcinoma (NPC). EBV persists asymptomatically in lifelong carriers due to a balance between its capability to transform B cells into proliferating lymphoblasts and a tight control of EBV+ B cells mainly by CD8+ T cells (1). Different forms of EBV latency are recognized, and these are defined by the extent of latent viral gene expression. On initial EBV infection, and in latently infected lymphoblastoid cell lines in culture, the full spectrum of EBV latency genes, EBV nuclear antigen 1 (EBNA1), EBNA2, EBNA3A, EBNA3B, EBNA3C, EBNA-LP, latent membrane protein 1 (LMP-1), LMP-2A, LMP-2B, and BamHI A rightward transcripts (BARTs), is expressed and this expression pattern has been termed latency III. EBV-associated tumors demonstrate a third pattern of latency-gene expression (latency I/II) in which only EBNA-1 and the BamHI-A rightward transcripts are expressed (latency I) or there is variable expression of the latency membrane proteins LMP-1, LMP-2A, and LMP-2B in addition to EBNA-1 and the BamHI-A rightward transcripts (latency II) (2-4).
Tumor cells in virus-associated tumors express viral antigens, and cytotoxic T lymphocytes (CTLs) responding to these antigens destroy the tumor cells (5). CTLs responding to EBV-specific proteins are induced by EBV infection or reactivation and these CTLs contribute to preventing EBV from proliferating within the body (6). In vitro CTL clonal expansion for BARTs suggests that EBV antigens with weak antigenicity could also induce EBV antigen-specific cell-mediated immunity (7). However, EBV antigen-specific cell-mediated immunity is prevented by either suppressing the expression of EBV antigens having strong antigenicity such as EBNA3A, and EBNA3C in EBV-associated tumors or self-inhibition of translation and antigen presentation by EBNA1 (8,9).
It was reported that EBV genomes are present and expressed in patients with gastric carcinoma (10,11). Despite the fact that only 6.9~16% of all gastric cancer cases are EBV positive, EBV is detected in more than 85% of lymphoepithelioma-like gastric carcinoma (LELC), in which lymphocyte infiltration is frequently seen (12,13). LELC whose lesions are mostly composed of lymphocytes is epithelial cell in origin but morphologically similar to lymphoma. LELC comprises 2% of all gastric cancer cases (14) Because the prognosis for LELC is good and most infiltrating lymphocytes are CD8+ T cells (15), it can be speculated that EBV specific proteins expressed in LELC would act as the antigens to induce the expansion of specific CTLs and that cell-mediated response by proliferated CTLs would affect tumor prognosis (16).
We investigated the expression of EBV genes in EBV-associated gastric carcinoma tissues and the presence of clonally expanded infiltrating T lymphocytes in cancer tissues by detecting the presence of the EBV genome and by measuring the TCR Vβ CDR3 usage in gastric cancer tissue samples obtained from 30 patients. We found expression of EBNA1 and BARTs in 4 cases. By measuring the length of the TCR Vβ CDR3 gene of CD8+ T lymphocytes that have infiltrated into gastric cancer tissues and by conducting sequence analysis of these genes, we also found the possibility that the infiltrating lymphocytes EBV-associated gastric carcinomas resulted from the clonally expanded CD8+ T lymphocyte population.
We examined 30 samples of primary gastric carcinoma resected from randomly selected Korean patients including 21 males and 9 females whose ages ranged from 34~76 years old. The positive and negative controls used for EBV gene expression were EBV positive B lymphoblastoid cell line and the gastric carcinoma cell line, MKN45, respectively. Cells were cultured in RPMI 1640 medium (Gibco BRL) containing 10% fetal bovine serum (Gibco BRL), 100 U/ml of penicillin and 100 µg/ml of streptomycin (Gibco BRL).
DNA PCR was performed using the primers SL18 (5'-GGCGCACCTGGAGGTGGTCC-3') and SL19 (5'-TTTCCAGCAGAGTCGCTAGG-3') to the variable number of tandem repeat (VNTR) in a mixture of 2 µl of 5 mM dNTP, 1.25 U Taq polymerase (Perkin Elmer, Branchburg, NJ) and 10 pmol primer set. PCR was done in a thermal cycler (Perkin Elmer) by carrying out the reaction in 35 cycles of 30 sec at 94℃, 30 sec at 55℃, and 30 sec at 72℃ with 10 min. extension at 72℃. The resulting PCR product was electrophoresed in 2% agarose gel and observed using an UV transilluminator after staining with ethidium bromide.
EBV-encoded small nonpolyadenylated RNAs (EBERs) constitutively expressed in EBV infected cells were used as the probe for in situ hybridization. Each of the 6 µm tissue samples was placed on a sialine coated slide, left for 30 min at 56℃, and placed in xylene twice for 3 min each to remove paraffin. The tissue sample was then placed twice in 99% and 95% ethyl alcohol solutions for 3 min each and hydrated by leaving the sample in distilled water. It was then reacted in Proteinase K (Sigma, St. Louis, MO) at 37℃ for 10 min, washed in distilled water, and completely dried by exposing to air for 20 min. After 40~100 µl FITC-labeled EBERs probe (Dako, Denmark) was placed on the dried tissue sample, it was reacted at 37℃ for 2 h. The sample was placed in tris buffered saline (0.1 M Tris in normal saline; TBS) for 5 min and washed 3 times in TBS containing 0.1% Triton X-100. After 100 µl anti-FITC (Dako) was placed onto the sample slide, the sample was reacted for 30 min at room temperature, washed with TBS, reacted with alkaline phosphatase-conjugated anti-mouse Ig for 30 min, and stained with BCIP/NBT.
In order to determine the type of EBV, PCR was performed using EBNA3C specific primers (sense: 5'-AGAAGGGGAGCGTGTGTTGT-3', antisense: 5'-GGCTCGTTTTTGACGTCGGC-3'). The EBV type was determined based on the fact that type 1 EBV would have an internal deletion at 93 bp, unlike type 2 EBV, so the PCR product of type 1 EBV would be 153 bp, while that of type 2 EBV would be 246 bp (17).
Reverse transcriptase-polymerase chain reaction (RT-PCR) was performed to determine the expression of EBV specific genes in gastric cancer tissues. RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA). A 30 µl reaction mixture was prepared by adding 5 µg RNA, 2 µg oligo-dT primer (Pharmacia, Uppsala, Sweden), 2 µl 10 mM dNTP (Boehringer Mannheim, Mannheim, Germany), 6 µl 5× reverse transcriptase buffer, 200 U M-MLV reverse transcriptase (Gibco BRL). cDNA was synthesized at 42℃ for 60 min. RT-PCR was performed using the synthesized cDNA as the template and the primer specific for each gene (18).
Immunohistochemistry was performed using anti-CD4, anti-CD8, and anti-CD20 antibodies to confirm the distribution of infiltrating T lymphocytes and B lymphocytes into gastric carcinoma in paraffin-embedded tissue sections. After leaving 6 µm of gastric carcinoma tissue sample on a sialine-coated slide, paraffin was removed. Add each primary antibody conjugated to alkaline phosphatase was placed onto the slide, the sample was reacted at 37℃ for 1 h and stained with BCIP/NBT.
For TCR Vβ CDR3 RT-PCR, the β chain constant region (Cβ: 5'-TTCTGATGGCTCAAACAC-3') was used as the antisense primer and a specific primer corresponding to each Vβ family was used as the sense primer (19,20). PCR was performed using 4 µl 1.25 mM dNTP, 1.25 U Taq polymerase (Perkin Elmer), and 10 pmol Vβ family specific primer and Cβ primer using cDNA synthesized from RNA extracted from gastric carcinoma tissues. PCR was performed by carrying out the reaction in 30 cycles of 30 sec at 94℃, 30 sec at 55℃, and 30 sec at 72℃, and extension was done at 72℃ for 10 min. After confirming the PCR product by electrophoresis in a 2% agarose gel, it was used as the nested-PCR template to determine the TCR Vβ family. Nested-PCR was performed using 1 µl TCR Vβ PCR product, 1 pmol sense Vβ primer, and 32P-labelled antisense Cβ primer. After electrophoresis of the PCR product in a sequencing gel, the gel was dried and exposed to Fuji Super HR-G X-ray film (Fuji Photo Film Co., Tokyo, Japan). The band intensity of each Vβ specific RT-PCR product was measured using the image plate and the predominant band was defined when the intensity of a single band was more than 1/3 of the total intensity. The predominant bands considered to be specific T lymphocyte clonal expansion were cut and extract DNA. PCR was performed with the extracted DNA using the same Vβ primer and Cβ primer, and the PCR products were confirmed using electrophoresis in a 2% agarose gel. PCR products of DNA extracted from the gel were cloned in pT7Blue-T vector (Novagen, La Jolla, CA), and the nucleotide sequences were analyzed. At the same time, direct sequencing analysis was done using the PCR product by using the AmpliCycle sequencing kit (Perkin Elmer). The nucleotide sequences of at least 4 of the clones that were cloned into the pT7 Blue-T vector were analyzed. Clonal expansion of a specific T lymphocyte was defined when the clone showing the same result as the direct sequencing of the PCR product was present in more than 2/3 of the total clones. The BLAST server by NCBI was used for Nucleotide database searching and computation.
PCR was performed using DNA extracted from gastric carcinoma tissue samples to confirm the presence of the EBV genome using EBV VNTR specific primers. The EBV genome was contained in gastric tissue samples obtained from 4 (#967, #974, #1010, and #1016) out of 30 patients (13.3%) (Fig. 1A). The fact that the VNTR PCR products of 4 tissue samples were seen as a single band (Fig. 1B) means that each case of gastric carcinoma was infected with a single EBV clonotype. As for VNTR, each EBV strain could have a different number of tandem repeats. Gastric carcinoma tissues with detected EBV genomes were undifferentiated adenocarcinomas according to H-E staining in which severe infiltration of lymphocytes was seen around the gastric carcinoma cells (Fig. 2A). Two (#967, #1016) of the 4 samples with the EBV genome were confirmed as LELC. According to the EBV EBER-1 in situ hybridization results, EBV genomes were observed in the nuclei of gastric carcinoma cells but not in the connective tissues and infiltrating lymphocytes (Fig. 2B).
To investigate the types of EBV infecting gastric carcinoma tissue samples with detectable EBV genomes, PCR was performed using EBNA3C specific primers. The EBNA3C gene shows the most significant difference between Type 1 and Type 2, in which an internal deletion is present in Type 1 EBV (17). The PCR product of Type 1 EBV was153 bp, and that of Type 2 EBV was 246 bp. The PCR results from all four samples showed the same size PCR product as that isolated from the B95.8 Type 1 EBV (Fig. 3), suggesting that all 4 samples were infected with Type 1 EBV.
To determine the type of latent infection present in gastric carcinoma tissues, which contain the EBV genome, the expression profile of EBV latent genes was investigated using RT-PCR. EBNA1, which is the most important for the maintenance of the EBV episome, was expressed in all 4 samples. The EBNA1, which is the gene that is expressed in all types of latent infection and that controls DNA replication of the EBV episome, was present in all samples. BARTs, which are usually expressed during EBV infection was detected in all 4 samples. LMP2a was expressed in one sample (#967) out of 4 samples. EBNA2 was not expressed in all 4 samples, suggesting that the C and W promoters were not activated. Because EBNA2, EBNA-LP, EBNA3A, EBNA3B, and EBNA3C expression is controlled by the C and W promoters, they were not expressed, either. LMP-1, expressed during Type 2 EBV latent infection, was not expressed (Fig. 4).
Immunohistochemistry was performed using anti-CD4, anti-CD8 and anti-CD20 antibodies to investigate the distribution of lymphocytes infiltrating EBV-positive gastric carcinoma tissue samples. Most lymphocytes were stained by anti-CD8 antibodies (Fig. 5A) but not with anti-CD4 and anti-CD20 antibodies (Fig. 5B and C), suggesting that most lymphocytes infiltrating gastric carcinoma tissues were CD8+ T cells.
Since the lymphocytes infiltrating gastric carcinoma tissues that contain the EBV genome were CD8+ T lymphocytes, TCR Vβ chain usage was analyzed to determine the presence of clonal expansion according to each Vβ family in order to determine whether single clonal or oligoclonal T lymphocytes recognizing a specific antigen were expanded. After performing RT-PCR with Vβ family specific primers, electrophoresis was done in a sequencing gel and CDR3 was sorted according to the different sizes (Fig. 6). After the band intensity of each RT-PCR product was measured according to each Vβ family, the predominant band was defined when the intensity of a single band was more than 1/3 of the total intensity. The predominant band was observed in Vβ family 2, 6 and 11 of T lymphocytes infiltrating sample #967; Vβ family 12 and 14 of sample #974; Vβ family 3, 6, 8, and 15 in sample #1010; and in Vβ family 8 of sample #1016. DNA was extracted from a total of 10 predominant bands and its nucleotide sequence was determined by using PCR-direct sequencing and also by cloning the PCR product with pT7 Blue-T vector at the same time. The two results were then compared. The nucleotide sequence of at least 4 clones that were cloned with pT7 Blue-T vector was analyzed. A specific T cell clone was expanded when the clone was more than 2/3 of the total clones showing the same result as the direct sequencing of the PCR product. Among the selected predominant bands, the nucleotide sequence obtained by direct sequencing was the same as the sequence obtained from the pT7 Blue-T vector with Vβ CDR3 cloned into it in all cases except for Vβ 12 of sample #974; Vβ 3, 6, and 15 of sample #1010; and Vβ 8 of sample #1016. The amino acid sequences deduced from these samples are shown in Table I. The length of CDR3 was determined by subtracting 4 amino acids from the number of amino acids present between the C-terminal cysteine in the Vβ region and the first glycine present on glycine/X/glycine in the J region. The CDR3 length was between 8 and 11, falling in the normal range of TCR Vβ CDR3. Its average was 9.6, which is similar to 9.5, the average length of TCR Vβ. When the Vβ family gene was searched using BLAST to confirm Jβ and C family, the result showed 3 clones using the same Jβ 2.3 TCR. Arginine was synthesized by N-addition in the V-D region of TCR Vβ of T lymphocytes infiltrating gastric carcinoma tissue samples #974 and #1010.
Gastric carcinoma is one of the most prevalent carcinomas in the world and is the most frequent carcinoma in Far East Asian countries such as Korea and Japan. Many studies reported various indirect evidences that EBV plays an important role in EBV-associated gastric carcinoma (21,22). Furthermore, EBV-associated gastric carcinoma, which has a good prognosis, shows some p53 mutations, which are frequently seen in stomach cancer (23), and severe infiltration of lymphocytes into tumor tissues (16,24). According to previous studies, EBV is crucial for malignant transformation in EBV-associated gastric carcinoma (25). At the same time, we could assume that EBV would enable the T lymphocytes of the host to recognize transformed tumor cells. Therefore, we determined the presence of EBV specific gene expression in EBV-associated gastric carcinoma and lymphocytes infiltrating into the tumor cells were clonal expansions of CD8+ T lymphocytes.
EBV-associated gastric carcinoma accounts for 6.9~16% of all cases of gastric carcinoma (10). We found the EBV genome in 4 (13.3%) of the 30 gastric carcinoma tissues samples. According to another study investigating the presence of the EBV genome in gastric carcinoma cases among Koreans, the EBV genome was detected in 13.5% of all gastric carcinoma cases (26). We found similar result in this study. According to histopathologic findings of gastric carcinoma tissue samples containing the EBV genome, the tissues were poorly differentiated and were severely infiltrated with lymphocytes. According to EBERs in situ hybridization, no EBV genome was detected in the surrounding lymphocytes. Thus, these results indicate that EBV was present in gastric carcinoma cells rather than in infected lymphocytes that infiltrated the gastric carcinoma. EBV can be largely divided into two types (Type 1 and Type 2), which have a slightly different gene composition (17). The distribution of these two types is similar in healthy individuals. However, most of the EBV isolated from tumors is Type 1, so that Type 1 EBV is more involved in tumorigenesis compared with Type 2 (27). EBV detected in all 4 samples in this study was also Type 1.
EBV is associated with many tumors and latent infection could be divided into 3 different types classified according to the specific genes expressed in the tumor cells (2). The expression of EBNA1, LMP-2 and BARTs were previously reported in EBV-associated gastric carcinoma (28). In this study, we observed the expression of only EBNA1, LMP-2 (1/4) and BARTs and no expression of EBNA2, suggesting that the C and W promoters are not activated. Furthermore, we did not observe the expression of LMP-1. Although EBNA1 expressed in Type 1 latent infection is expected to play an essential role in malignant cell transformation, the mechanism involved in this process has not been elucidated.
According to immunohistochemistry and sequence analysis of the TCR Vβ chain of CDR3, we found that there was an expansion of 5 specific T lymphocyte clones. It was likely that these clones were expanded tumor-specific CD8+ T lymphocytes, suggesting that tumor specific antigens are present in EBV-associated gastric carcinoma and that CD8+ T lymphocytes responding to these antigens have infiltrated tumor tissues. Thus, the good prognosis of EBV-associated gastric carcinomas could be accounted for by these CD8+ T lymphocytes that result in tumor-specific cytotoxicity. The fact that we could not observe the same TCR clonal expansion in 3 samples suggests that CD8+ T lymphocyte clonal expansion did not respond to superantigens. This is in contrast to results reported previously that lytic cycle specific proteins would function as superantigens to expand CTL clone at the time of primary EBV infection (29). We observed various CD8+ T lymphocyte clonal expansions possibly because antigens expressed during latent infection are conventional antigens and specific CD8+ T lymphocytes would recognize various antigenic peptides originating from EBV proteins that form complexes with MHC class I molecules. In this study, we found that CD8+ T lymphocytes infiltrated EBV-associated gastric carcinoma cells and that EBNA1, LMP2a and BARTs were expressed. This suggests that EBNA1, LMP2, or BARTs could provide the antigens that result in the expansion of CD8+ T lymphocytes. Since EBNA1 has the Gly-Ala rich domain that prevents antigen processing and presentation, EBNA1 is not immunogenic to Human CD8+ T cell (30). However, it was reported that Human CD4+ T cells consistently respond to the EBNA1, and BARTs LMP2a could act as an antigenic determinant in NPC and HD (7,31,32).
We detected the presence of the EBV genome, specifically Type 1 EBV, in gastric carcinoma tissues. We observed the expression of EBNA1, LMP2a and BARTs in gastric carcinoma tissue samples that have the EBV genome but found no expression of EBNA2 and LMP-1. We found that lymphocytes infiltrating in gastric carcinoma tissues in EBV-associated gastric carcinoma were CD8+ T lymphocyte clones responding to specific conventional antigens. We determined this by measuring the length and analyzing the nucleotide sequence of TCR Vβ CDR3 of T lymphocytes that have infiltrated gastric carcinoma tissues. This result suggests that EBV-specific proteins act as antigens in EBV-associated gastric carcinoma to induce a cell-mediated immune response. Further in vitro CTL proliferation studies using EBV-specific proteins are needed to determine specific epitopes.
Figures and Tables
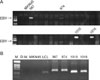 | Figure 1Detection of EBV DNA in gastric carcinoma tissues using PCR. (A) DNA was extracted from 30 gastric carcinoma tissue samples, and PCR was performed using the EBV specific primers SL18 and SL19. PCR products were electrophoresed in a 2% agarose gel. The EBV genome was detected in 4 of the samples (#967, #974, #1010, and #1016). No EBV genome was detected in the gastric carcinoma cell line, MKN45 (negative control), and 26 gastric carcinoma tissue samples. Lane 1: DNA size marker, 1 kb DNA ladder; MKN45: gastric carcinoma cell line showing no EBV infection; 967, 974, 1010, 1016: Gastric carcinoma tissues samples with the EBV genome. (B) VNTR DNA of EBV LMP1 was amplified and electrophoresis was performed. M: DNA size marker, 100 bp DNA ladder; DW: distilled water; MKN45: gastric carcinoma cell line (negative control); LCL: B lymphoblastoid cell line (positive control) transformed with EBV produced with the B95.8 cell line; 967, 974, 1010, 1016: gastric carcinoma tissue samples with EBV genes detected. |
 | Figure 2Pathological findings of gastric carcinoma tissue samples and EBER in situ hybridization. (A) The pathological findings of gastric carcinoma tissue sample #967 containing the EBV genome. This carcinoma was an undifferentiated adenocarcinoma and shows severe infiltration of lymphocytes (H-E staining, ×400 magnification). (B) EBERs in situ hybridization. Nuclei of gastric carcinoma cells show a positive response, while lymphocytes infiltrating the gastric carcinoma tissues and the surrounding connective tissue show a negative response. |
 | Figure 3Determination of EBV type in gastric carcinoma tissues. PCR was performed using EBNA3C specific primers in order to determine the type of EBV in gastric carcinoma tissues. The resulting PCR product was electrophoresed. Lane 1: DNA size marker, 100 bp DNA ladder; MKN45: negative control; B95.8: positive control; 967, 974, 1010, and 1016: gastric carcinoma tissue samples with EBV genes detected. |
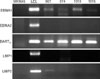 | Figure 4Detection of EBV specific gene transcripts in gastric carcinoma tissue samples using RT-PCR. Total RNA was isolated from gastric carcinoma tissues and cDNA was prepared. RT-PCR was performed using the cDNA as template and specific primers for EBV genes, i.e., EBNA1, EBNA2, BARTs and LMP-1. The resulting products were electrophoresed in a 2% agarose gel. MKN45: negative control; LCL: B lymphoblastoid cell line (positive control) transformed with EBV produced with the B95.8 cell line; 967, 974, 1010, 1016: gastric carcinoma tissue samples with EBV gene detected. |
 | Figure 5Distribution of lymphocytes infiltrating in gastric carcinoma tissues. Immunohistochemical staining of gastric carcinoma tissue #967 using anti-CD8 (A), anti-CD4 (B), and anti-CD20 (C) antibodies (200×). |
 | Figure 6TCR Vβ chain usage of DR3 of CD8+ T cells infiltrating gastric carcinoma tissues. After performing RT-PCR using specific primers to each Vβ family, electrophoresis was done in a sequencing gel and CDR3 was divided according to sizes. The predominant band (arrow) could be observed in Vβ family 2, 6, and 11 of sample #967; Vβ family 12 and 14 of sample #974; Vβ family 3, 6, 8, and 15 of sample #1010;Vβ family 8 of sample #1016. 967, 974, 1010, and 1016: Gastric carcinoma tissue samples with EBV gene detected. |
ACKNOWLEDGEMENTS
We thank In Hong Choi and Yong-Joon Chwae for some of the reagents used in these studies. We thank Kim, Young-Mi for excellent technical assistance.
This study was supported by a faculty research grant of Yonsei University College of Medicine for 2004 (6-2004-1079).
References
1. Rickinson AB, Kieff E. Knipe DM, Howley PM. Epstein-Barr virus. Fields virology. 2007. 5th ed. Philadelphia: Lippincott Williams & Wilkins;2655–2700.
2. Kieff E, Rickinson AB. Fields BN, Knipe DM, Howley PM. Epstein-Barr Virus and its replication. Fields Virology. 2007. 5th ed. Philadelphia: Lippincott-Williams & Wilkins Publishers;2603–2654.
3. Sample J, Brooks L, Sample C, Young L, Rowe M, Gregory C, Rickinson A, Kieff E. Restricted Epstein-Barr virus protein expression in Burkitt lymphoma is due to a different Epstein-Barr nuclear antigen 1 transcriptional initiation site. Proc Natl Acad Sci U S A. 1991. 88:6343–6347.

4. Young LS, Murray PG. Epstein-Barr virus and oncogenesis: from latent genes to tumours. Oncogene. 2003. 22:5108–5121.

5. Boon T, van der Bruggen P. Human tumor antigens recognized by T lymphocytes. J Exp Med. 1996. 183:725–729.

6. Rickinson AB, Moss DJ. Human cytotoxic T lymphocyte responses to Epstein-Barr virus infection. Annu Rev Immunol. 1997. 15:405–431.

7. Kienzle N, Sculley TB, Poulsen L, Buck M, Cross S, Raab-Traub N, Khanna R. Identification of a cytotoxic T-lymphocyte response to the novel BARF0 protein of Epstein-Barr virus: a critical role for antigen expression. J Virol. 1998. 72:6614–6620.

8. Blake N, Lee S, Redchenko I, Thomas W, Steven N, Leese A, Steigerwald-Mullen P, Kurilla MG, Frappier L, Rickinson A. Human CD8+ T cell responses to EBV EBNA1: HLA class I presentation of the (Gly-Ala)-containing protein requires exogenous processing. Immunity. 1997. 7:791–802.

9. Yin Y, Manoury B, Fåhraeus R. Self-inhibition of synthesis and antigen presentation by Epstein-Barr virus-encoded EBNA1. Science. 2003. 301:1371–1374.

10. Akiba S, Koriyama C, Herrera-Goepfert R, Eizuru Y. Epstein-Barr virus associated gastric carcinoma: epidemiological and clinicopathological features. Cancer Sci. 2008. 99:195–201.

11. Burke AP, Yen TS, Shekitka KM, Sobin LH. Lymphoepithelial carcinoma of the stomach with Epstein-Barr virus demonstrated by polymerase chain reaction. Mod Pathol. 1990. 3:377–380.
12. Herath CH, Chetty R. Epstein-Barr virus-associated lymphoepithelioma-like gastric carcinoma. Arch Pathol Lab Med. 2008. 132:706–709.

13. Nakamura S, Ueki T, Yao T, Ueyama T, Tsuneyoshi M. Epstein-Barr virus in gastric carcinoma with lymphoid stroma. Special reference to its detection by the polymerase chain reaction and in situ hybridization in 99 tumors, including a morphologic analysis. Cancer. 1994. 73:2239–2249.

14. Corvalan A, Ding S, Koriyama C, Carrascal E, Carrasquilla G, Backhouse C, Urzua L, Argandoña J, Palma M, Eizuru Y, Akiba S. Association of a distinctive strain of Epstein-Barr virus with gastric cancer. Int J Cancer. 2006. 118:1736–1742.

15. Minamoto T, Mai M, Watanabe K, Ooi A, Kitamura T, Takahashi Y, Ueda H, Ogino T, Nakanishi I. Medullary carcinoma with lymphocytic infiltration of the stomach. Clinicopathologic study of 27 cases and immunohistochemical analysis of the subpopulations of infiltrating lymphocytes in the tumor. Cancer. 1990. 66:945–952.

16. van Beek J, zur Hausen A, Snel SN, Berkhof J, Kranenbarg EK, van de Velde CJ, van den Brule AJ, Middeldorp JM, Meijer CJ, Bloemena E. Morphological evidence of an activated cytotoxic T-cell infiltrate in EBV-positive gastric carcinoma preventing lymph node metastases. Am J Surg Pathol. 2006. 30:59–65.

17. Sample J, Young L, Martin B, Chatman T, Kieff E, Rickinson A, Kieff E. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol. 1990. 64:4084–4092.

18. Sugiura M, Imai S, Tokunaga M, Koizumi S, Uchizawa M, Okamoto K, Osato T. Transcriptional analysis of Epstein-Barr virus gene expression in EBV-positive gastric carcinoma: unique viral latency in the tumour cells. Br J Cancer. 1996. 74:625–631.

19. Choi IH, Chwae YJ, Shim WS, Kim DS, Kwon DH, Kim JD, Kim SJ. Clonal expansion of CD8+ T cells in Kawasaki disease. J Immunol. 1997. 159:481–486.
20. Hall BL, Finn OJ. PCR-based analysis of the T-cell receptor V beta multigene family: experimental parameters affecting its validity. Biotechniques. 1992. 13:248–257.
21. Shibata D, Weiss LM. Epstein-Barr virus-associated gastric adenocarcinoma. Am J Pathol. 1992. 140:769–774.
23. Gulley ML, Pulitzer DR, Eagan PA, Schneider BG. Epstein-Barr virus infection is an early event in gastric carcinogenesis and is independent of bcl-2 expression and p53 accumulation. Hum Pathol. 1996. 27:20–27.

24. van Beek J, zur Hausen A, Klein Kranenbarg E, van de Velde CJ, Middeldorp JM, van den Brule AJ, Meijer CJ, Bloemena E. EBV-positive gastric adenocarcinomas: a distinct clinicopathologic entity with a low frequency of lymph node involvement. J Clin Oncol. 2004. 22:664–670.

25. Imai S, Koizumi S, Sugiura M, Tokunaga M, Uemura Y, Yamamoto N, Tanaka S, Sato E, Osato T. Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc Natl Acad Sci U S A. 1994. 91:9131–9135.

26. Shin WS, Kang MW, Kang JH, Choi MK, Ahn BM, Kim JK, Sun HS, Min KW. Epstein-Barr virus-associated gastric adenocarcinomas among Koreans. Am J Clin Pathol. 1996. 105:174–181.

27. Sixbey JW, Shirley P, Chesney PJ, Buntin DM, Resnick L. Detection of a second widespread strain of Epstein-Barr virus. Lancet. 1989. 2:761–765.

29. Steven NM, Annels NE, Kumar A, Leese AM, Kurilla MG, Rickinson AB. Immediate early and early lytic cycle proteins are frequent targets of the Epstein-Barr virus-induced cytotoxic T cell response. J Exp Med. 1997. 185:1605–1617.

30. Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen PM, Klein G, Kurilla MG, Masucci MG. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995. 375:685–688.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download