1). Burnett AL. Strategies to promote recovery of cavernous nerve function after radical prostatectomy. World J Urol. 2003; 20:337–42.

2). Olsson CA, Goluboff ET. Detection and treatment of prostate cancer: perspective of the urologist. J Urol. 1994; 152:1695–9.

3). Walsh PC, Donker PJ. Impotence following radical prostatectomy: insight into etiology and prevention. J Urol. 1982; 128:492–7.

4). Takenaka A, Murakami G, Matsubara A, Han SH, Fujisawa M. Variation in course of cavernous nerve with special reference to details of topographic relationships near prostatic apex: histologic study using male cadavers. Urology. 2005; 65:136–42.

5). Lepor H, Gregerman M, Crosby R, Mostofi FK, Walsh PC. Precise localization of the autonomic nerves from the pelvic plexus to the corpora cavernosa: a detailed anatomical study of the adult male pelvis. J Urol. 1985; 133:207–12.

6). Montorsi F, Briganti A, Salonia A, Rigatti P, Burnett AL. Current and future strategies for preventing and managing erectile dysfunction following radical prostatectomy. Eur Urol. 2004; 45:123–33.

7). Costello AJ, Brooks M, Cole OJ. Anatomical studies of the neurovascular bundle and cavernosal nerves. BJU Int. 2004; 94:1071–6.

8). Sezen SF, Burnett AL. Intracavernosal pressure monitoring in mice: responses to electrical stimulation of the cavernous nerve and to intracavernosal drug administration. J Androl. 2000; 21:311–5.
9). Lysiak JJ, Yang SK, Klausner AP, Son H, Tuttle JB, Steers WD. Tadalafil increases Akt and extracellular signal-regulated kinase 1/2 activation, and prevents apoptotic cell death in the penis following denervation. J Urol. 2008; 179:779–85.

10). Mullerad M, Donohue JF, Li PS, Scardino PT, Mulhall JP. Functional sequelae of cavernous nerve injury in the rat: is there model dependency. J Sex Med. 2006; 3:77–83.

11). Klein LT, Miller MI, Buttyan R, Raffo AJ, Burchard M, Devris G, et al. Apoptosis in the rat penis after penile denervation. J Urol. 1997; 158:626–30.

12). User HM, Hairston JH, Zelner DJ, McKenna KE, McVary KT. Penile weight and cell subtype specific changes in a post-radical prostatectomy model of erectile dysfunction. J Urol. 2003; 169:1175–9.

13). Hu WL, Hu LQ, Song J, Li SW, Zheng XM, Cheng B, et al. Fibrosis of corpus cavernosum in animals following cavernous nerve ablation. Asian J Androl. 2004; 6:111–6.
14). Carrier S, Zvara P, Nunes L, Kour NW, Rehman J, Lue TF. Regeneration of nitric oxide synthase-containing nerves after cavernous nerve neurotomy in the rat. J Urol. 1995; 153:1722–7.

15). Iacono F, Giannella R, Somma P, Manno G, Fusco F, Mirone V. Histological alterations in cavernous tissue after radical prostatectomy. J Urol. 2005; 173:1673–6.

16). Domes T, De Young L, O'Gorman DB, Gan BS, Bella AJ, Brock G. Is there a role for proteomics in Peyronie's disease? J Sex Med. 2007; 4:867–77.

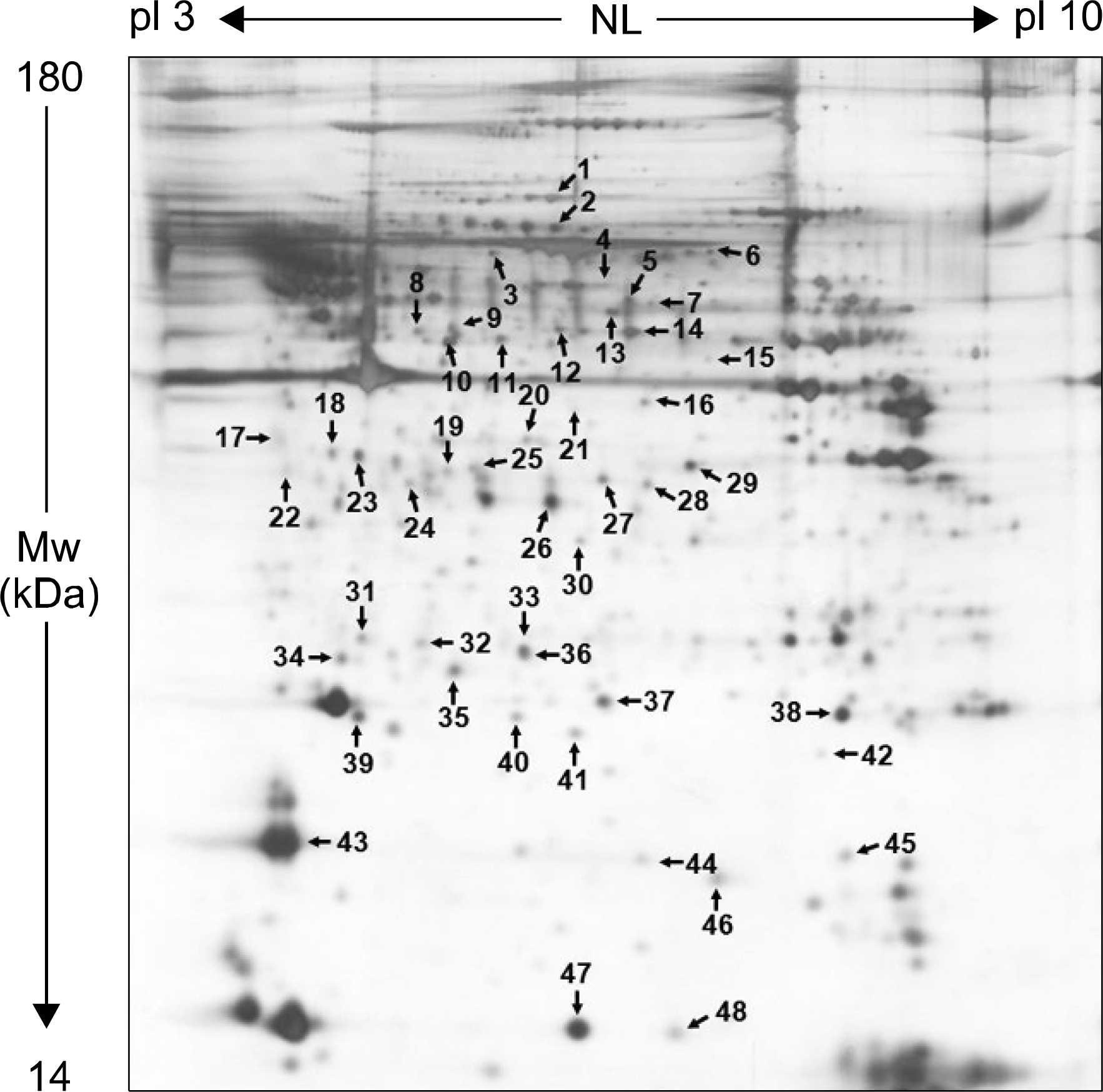
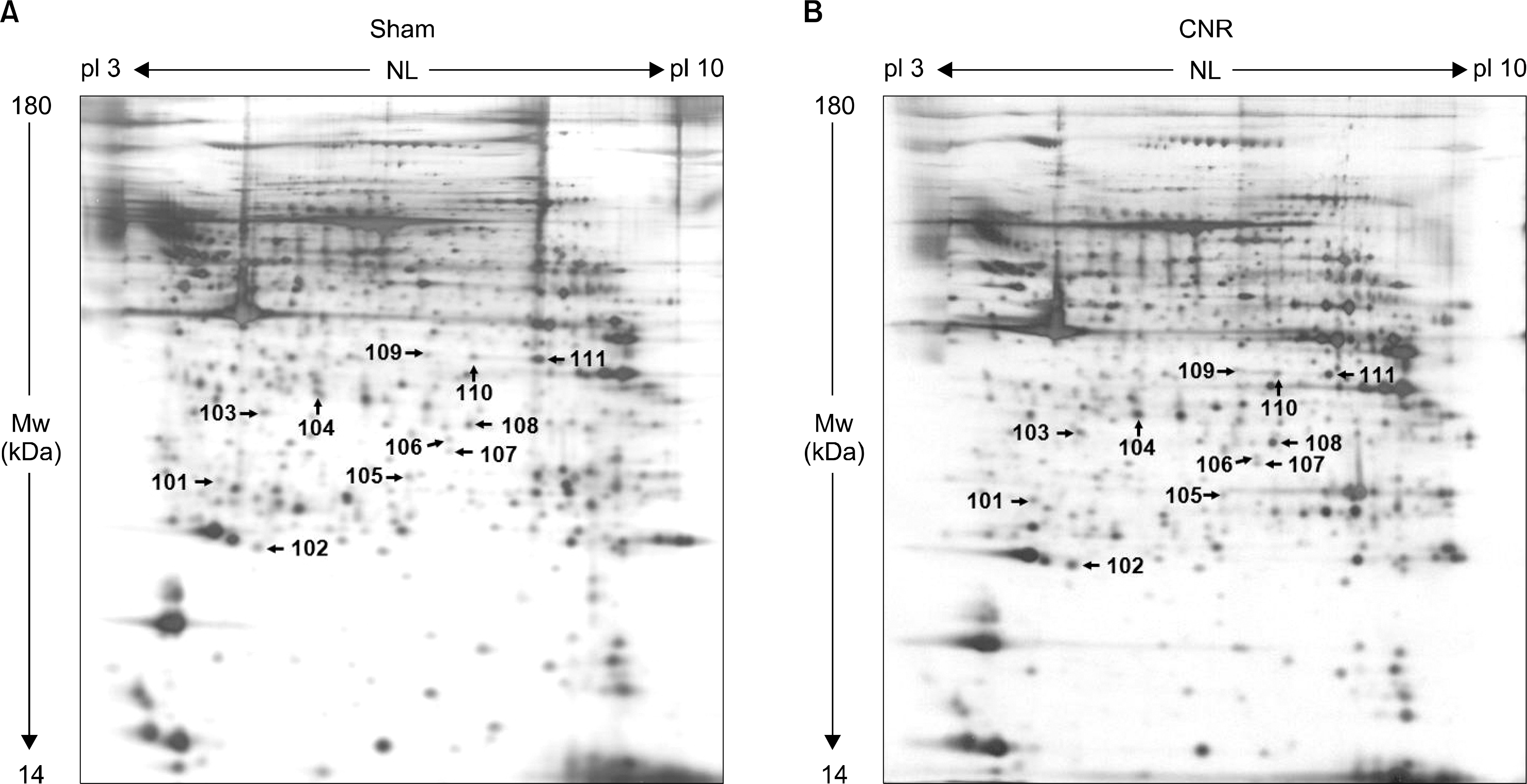
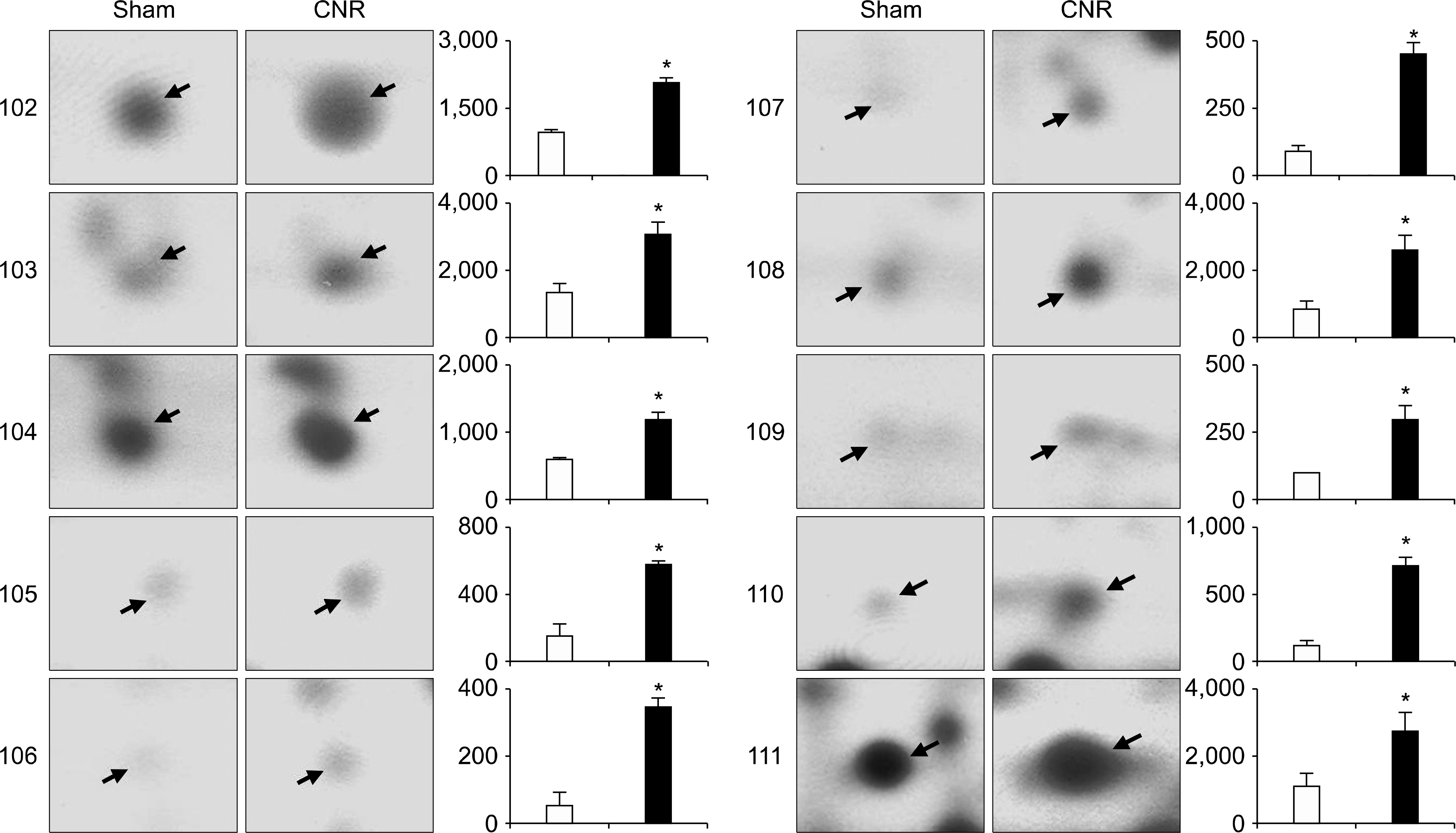
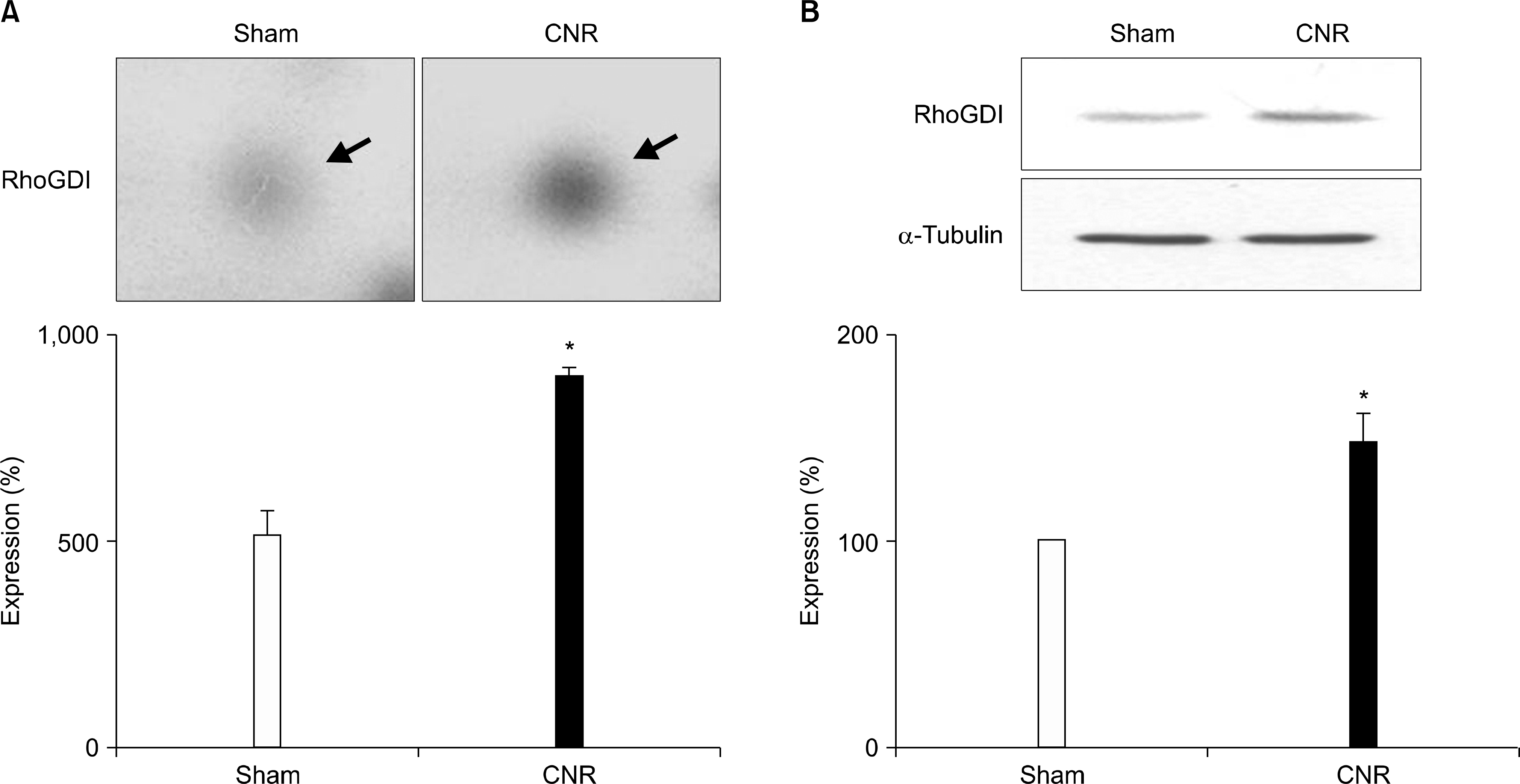
17). Liu X, Gao X, Pang J, Zhang Y, Wang K, Fang Y, et al. Proteomic analysis of rat penile tissue in a model of erectile dysfunction after radical prostatectomy. BJU Int. 2007; 99:1500–5.

18). User HM, Zelner DJ, McKenna KE, McVary KT. Microarray analysis and description of SMR1 gene in rat penis in a post-radical prostatectomy model of erectile dysfunction. J Urol. 2003; 170:298–301.

19). Lee CK, Park HJ, So HH, Kim HJ, Lee KS, Choi WS, et al. Proteomic profiling and identification of cofilin responding to oxidative stress in vascular smooth muscle. Proteomics. 2006; 6:6455–75.

20). Lee CK, Han JS, Won KJ, Jung SH, Park HJ, Lee HM, et al. Diminished expression of dihydropteridine reductase is a potent biomarker for hypertensive vessels. Proteomics. 2009; 9:4851–8.

21). Tostes RC, Carneiro FS, Lee AJ, Giachini FR, Leite R, Osawa Y, et al. Cigarette smoking and erectile dysfunction: focus on NO bioavailability and ROS generation. J Sex Med. 2008; 5:1284–95.

22). Wolf BB, Green DR. Suicidal tendencies: apoptotic cell death by caspase family proteinases. J Biol Chem. 1999; 274:20049–52.

23). Sasaki T, Takai Y. The Rho small G protein family-Rho GDI system as a temporal and spatial determinant for cytoskeletal control. Biochem Biophys Res Commun. 1998; 245:641–5.

24). Hoffman GR, Nassar N, Cerione RA. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell. 2000; 100:345–56.

25). Miñambres R, Guasch RM, Perez-Aragó A, Guerri C. The RhoA/ROCK-I/MLC pathway is involved in the ethanol-induced apoptosis by anoikis in astrocytes. J Cell Sci. 2006; 119:271–82.
26). Harenberg A, Girkontaite I, Giehl K, Fischer KD. The Lsc RhoGEF mediates signaling from thromboxane A2 to actin polymerization and apoptosis in thymocytes. Eur J Immunol. 2005; 35:1977–86.

27). He H, Baldwin GS. Rho GTPases and p21-activated kinase in the regulation of proliferation and apoptosis by gastrins. Int J Biochem Cell Biol. 2008; 40:2018–22.

28). Alici B, Gümüstas MK, Ozkara H, Akkus E, Demirel G, Yencilek F, et al. Apoptosis in the erectile tissues of diabetic and healthy rats. BJU Int. 2000; 85:326–9.

29). Jin L, Liu T, Lagoda GA, Champion HC, Bivalacqua TJ, Burnett AL. Elevated RhoA/Rho-kinase activity in the aged rat penis: mechanism for age-associated erectile dysfunction. FASEB J. 2006; 20:536–8.

30). Wang H, Eto M, Steers WD, Somlyo AP, Somlyo AV. RhoA-mediated Ca2+ sensitization in erectile function. J Biol Chem. 2002; 277:30614–21.
31). Sakamoto T, Repasky WT, Uchida K, Hirata A, Hirata F. Modulation of cell death pathways to apoptosis and necrosis of H2O2-treated rat thymocytes by lipocortin I. Biochem Biophys Res Commun. 1996; 220:643–7.

32). Parente L, Solito E. Annexin 1: more than an anti-phospholipase protein. Inflamm Res. 2004; 53:125–32.

33). Townsend DM, Tew KD. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene. 2003; 22:7369–75.

34). Bambang IF, Xu S, Zhou J, Salto-Tellez M, Sethi SK, Zhang D. Overexpression of endoplasmic reticulum protein 29 regulates mesenchymal-epithelial transition and suppresses xenograft tumor growth of invasive breast cancer cells. Lab Invest. 2009; 89:1229–42.






 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download