Abstract
Natural Killer (NK) cells are the third population of lymphocyte in the mononuclear cell compartment that triggers first-line of defense against viral infection and tumor transformation. Historically, NK cells were thought of as components of innate immunity based on their intrinsic ability to spontaneously kill target cells independent of HLA antigen restriction. However, it is now clear that NK cells are quite sophisticated and use a highly specific and complex target cell recognition receptor system arbitrated via a multitude of inhibitory and activating receptors. Killer cell immunoglobulin-like receptors (KIR) are the key receptors of human NK cells development and function. To date, fourteen distinct KIRs have been identified: eight are inhibitory types, and six are activating types. The number and type of KIR genes present varies substantially between individuals. Inhibitory KIRs recognize distinct motifs of polymorphic HLA class I molecules. Upon engagement of their specific HLA class I ligands, inhibitory KIR dampen NK cell reactivity. In contrast, activating KIRs are believed to stimulate NK cell reactivity when they sense their ligands (unknown). KIR and HLA gene families map to different human chromosomes (19 and 6, respectively), and their independent segregation produces a wide diversity in the number and type of inherited KIR-HLA combinations, likely contributing to overall immune competency. Consistent with this hypothesis, certain combinations of KIR-HLA variants have been correlated with susceptibility to diseases as diverse as autoimmunity, viral infections, and cancer. This review summarizes our emerging understanding of KIR-HLA diversity in human health and disease.
Natural killer (NK) cells of innate immunity and CD8+ cytotoxic T cells (CTL) of adaptive immunity are professional killer cells that are necessary for defense against virus-infected or tumor-transformed cells. NK cells and CTLs both originate from a common lymphoid progenitor and share several features in their development, morphology, cell-surface phenotypes and effector functions [1]. Recent reports further reveal unexpected similarities between these killer subsets in developing specific memory response that repeal the fundamental discrepancy between innate and adaptive immune cells [2, 3]. Similar to T and B cells of adaptive immunity, NK cells are educated during development, possess antigen-specific receptors, undergo clonal expansion during infection and generate long-lived memory cells. However, NK cells and CTLs differ in their cell surface receptors used to distinguish unhealthy cellular targets from the healthy host cells (Fig. 1). Whereas CTL activation is triggered through the interaction of a somatically re-arranged T cell receptor with a specific foreign peptide-laden human leukocyte antigen (HLA) class I molecule displayed by infected cells, the NK cells use a multitude of germ line-encoded receptor recognition strategies, whereby an individual NK cell can be triggered through various receptors with inhibitory or activating function, independently or in combination, depending on the ligands presented by the target cell in a given encounter [4-6]. If a given NK cell uses both inhibitory and activating receptors to recognize the target, the balance between these disparate signals determines the action of that NK cell [7].
Several gene families encode receptors for NK cells that trigger inhibitory or activating function, such as (i) killer cell immunoglobulin-like receptors (KIR), (ii) killer cell lectin-like receptors (KLR), (iii) leukocyte immunoglobulin-like receptors (LILR), and (iv) natural cytotoxicity receptors (NCR) [7-9]. Most of these receptors are expressed in a stochastic manner such that each NK cell clone within a given individual does not express the entire set of NK receptor genes present in that individual's genome, but rather only a portion of the genes in an apparently random combination [6, 10]. Variegated expression of multiple activating and inhibitory receptors results in an unexpected heterogeneity in NK subsets that yields a broad range of functionally distinct NK cell clones, which presumably are critical for a rapid and sensitive detection of infected cellular targets.
Because NK cells circulate in a state that can spontaneously deliver effector function, it is essential to block them from attacking healthy tissues. NK cells in peripheral blood express at least one inhibitory receptor for self-HLA class I molecules and thus provide tolerance to healthy cells that normally express abundant HLA class I molecules [6]. Down regulation of HLA class I expression due to certain viral infections, neoplastic transformations, or other forms of stress, relieves the inhibitory influence on NK cells, permitting NK cells to eliminate these unhealthy cells, a phenomenon originally described as the 'missing-self' hypothesis [11] (Fig. 1). Furthermore, by recognizing unknown ligands expressed on the target cells, such as 'induced-self' that are structurally related to HLA class I molecules (e.g., MICA and MICB), 'altered-self' (HLA class I molecule loaded with foreign peptide) or pathogen encoded 'non-self' (molecules associated with infection and tumor transformation), the activating receptors may further augment NK lysis of unhealthy cells.
Originating in simian primates, the KIR is relatively a new gene family that evolves rapidly to keep up with species-specific evolution and adaptation [12]. Because KIRs have considerable species-specificity and are lacking in rodents, it is critical to study humans to understand the impact of KIR on health and disease. KIR receptors play a key role in human NK cell development and function, and differ from other NK cell receptors by their substantial diversity, which is contributed by individual-specific KIR gene content and nucleotide sequence polymorphism of each KIR gene. Fourteen KIR receptors (plus 2 pseudogenes; 2DP1 and 3DP1) have been identified in humans that trigger inhibition (3DL1-3, 2DL1-3, 2DL5), activation (3DS1, 2DS1-5), or both (2DL4) (Fig. 2).
KIR receptors are encoded by a family of tightly clustered genes on the leukocyte receptor complex (LRC) spanning a region of about 150 kb on chromosome 19q13.4 [13]. Being diploid, humans ordinarily have two copies of each autosomal gene, one per chromosome. However, the KIR gene family violates this basic rule due to deletion or duplication. The number and type of KIR genes may differ substantially between two chromosomes (Fig. 3). Over 30 distinct KIR haplotypes with distinct gene content have been characterized to date by sequencing genomic clones and haplotype segregation analysis in families [13-18]. One of the haplotypes that occurs in all populations is conventionally named the 'A haplotype', which consists of nine genes (3DL3-2DL3-2DP1-2DL1-3DP1-2DL4-3DL1-2DS4-3DL2) and encodes predominantly inhibitory receptors (Fig. 3; haplotype 1) [13]. Remaining haplotypes are collectively referred to as 'group B haplotypes', which have variable gene content comprising several genes (2DS1, 2DS2, 2DS3, 2DS5, 2DL2, 2DL5, and 3DS1) that are not part of the A haplotype, and thus B haplotypes encode more activating KIRs compared with the A haplotype that encodes a single activating receptor, KIR2DS4 (Fig. 3; haplotypes 2-22). In addition to the substantial variation in gene content across haplotypes, each KIR gene itself exhibits considerable nucleotide sequence polymorphism [18-20]. To date, 614 KIR nucleotide sequences encoding 321 distinct KIR proteins (Fig. 2) have been deposited in IPD-KIR database (Release 2.4.0, 15 April 2011), a database that provides the centralized repository for human KIR sequences (http://www.ebi.ac.uk/ipd/kir/). The sequence polymorphism can influence the expression, ligand binding, and functional capacity [16, 21-24]. A large number of amino acid substitutions occurred among the allelic variants of each inhibitory KIR gene are predominantly located at the sites not directly implicated in HLA class I ligand binding [25, 26]. A number of these substitutions were shown to be the subject of positive selection, and thus the evolutionary pressure that drives KIR sequence polymorphism is presumably more than polymorphic HLA class I recognition and possibly involves rapidly evolving pathogen recognition.
Only four KIR genes (2DL4, 3DL2, 3DL3, and 3DP1) are present on all haplotypes and are referred to as 'framework' genes, while the existence of 12 other genes is considerably variable by haplotype [13]. A stretch of 14 kb DNA enriched with L1 repeats placed between 3DP1 and 2DL4 divides the KIR haplotype into two halves [13] (Fig. 3). The centromeric half is delimited by 3DL3 at the 5'-end and 3DP1 at the 3'-end, while the telomeric half is delimited by 2DL4 at the 5'-end and 3DL2 at the 3'-end. The inhibitory receptors, KIR2DL2 and 2DL3, segregate as alleles of a single locus at the centromeric half. Similarly, the inhibitory KIR3DL1 and activating KIR3DS1 behave as alleles of the same locus at the telomeric half. Almost all haplotypes contain these two loci, such that virtually everyone has either 2DL2 or 2DL3, and 3DL1 or 3DS1 within their KIR genome. KIR2DL1, 2DL2, 2DL3 and 2DS2 are specific to the centromeric half, while KIR3DL1, 3DS1, 2DS1, and 2DS4 are specific to the telomeric half. Three KIR genes, 2DL5, 2DS3 and 2DS5, are found in both centromeric and telomeric locations [15, 27]. For genes within each half, there is significant linkage disequilibrium, but much less for genes in the two different halves [18, 19].
Multiple reciprocal recombination events at the center of the KIR complex, between 3DP1 and 2DL4, presumably diversify gene content for KIR haplotypes across individuals and populations [28, 29]. Most of the KIR gene content haplotypes published to date can be explained by the recombination of 10 centromeric and 10 telomeric gene content motifs (Fig. 4). For example, haplotype 1 listed in Fig. 3 is a recombinant of the centromeric half cA01 and telomeric half tA01, similarly haplotype 8 is a recombinant of the centromeric half cB02 and telomeric half tB02. The reciprocal recombination also results in haplotypes carrying both group-A and group-B haplotype-specific motifs. For instance, haplotype 6 listed in Fig. 3 is a recombinant of the centromeric half cA01 and telomeric half tB04, and haplotype 9 is a recombinant of the centromeric half cB03 and telomeric tA01. Recombination events also are reported outside the region between 3DP1 and 2DL4. These are generally non-allelic crossovers generating several unusual haplotypes, including truncated haplotypes that are missing some framework genes [15, 19] or elongated haplotypes that contain duplicated genes [30].
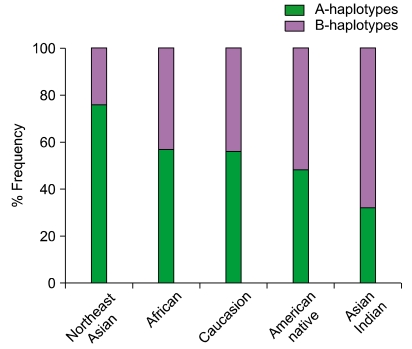
Inheritance of paternal and maternal haplotypes comprising different gene content (A+A, A+B, or B+B) generates extraordinary KIR diversity in humans (Fig. 5). For example, carriers of two A haplotypes have only seven functional KIR genes, while the carriers of both A and B haplotypes may have all 14 functional KIR genes. Several population studies revealed over 300 genotypes that differ in their KIR gene content and each population carries a distinct gene content profile [28, 31] (http://www.allelefrequencies.net/). All human populations have both group-A and -B haplotypes, but their distribution varies considerably across distinct populations (Fig. 6). Individuals carrying both A and B haplotypes are more common in Caucasians and Africans, and therefore A and B haplotypes are approximately equally distributed in these populations [28, 32]. In contrast, the prevalence of A haplotypes dominates over the B haplotypes in one population and vice versa in others. Individuals carrying homozygous A haplotypes (AA genotypes) are common in Northeast Asians (Chinese, Japanese, and Koreans) [17, 33, 34]. Conversely, individuals carrying Bx genotypes (either AB or BB) are common in the natives of America [35, 36], Australia [37], and India [38-40]. The NK cells from AA genotype carriers can express a maximum of four inhibitory KIRs (2DL1, 2DL3, 3DL1 and 3DL2) and one activating KIR (2DS4). In contrast, individuals carrying Bx genotypes can express a maximum of six inhibitory KIRs (2DL1-3, 2DL5, 3DL1, and 3DL2) and 2 to 6 activating KIRs (3DS1, 2DS1-5). NK cells of Bx genotype carriers express more activating KIR receptors and respond more vigorously to a range of pathogens. These data suggest that the aboriginal populations of India, Australia, and America acquired activating KIR genes to survive disease epidemics and population bottlenecks during their extensive pre-historic migrations from Africa [38].
KIR receptors recognize specific motifs of HLA class I molecules, which are the products of highly polymorphic genes of the major histocompatibility complex (MHC) located on chromosome 6 [41-43] (Fig. 2). HLA-C is the dominant HLA class I locus that provides ligands for many KIR receptors. All HLA-C allotypes carry valine (V) at position 76, while position 80 displays a dimorphism of either asparagine (N) or lysine (K). Nearly half of the HLA-C allotypes (Cw2, Cw4, Cw5, Cw6, and Cw15) carry a K80 (conventionally termed C2 epitope) that binds inhibitory receptor KIR2DL1 [44-46]. The remaining HLA-C allotypes (Cw1, Cw3, Cw7, and Cw8) carry N80 (termed C1 epitope) and bind inhibitory receptors KIR2DL2 and 2DL3. Two exceptionally diverse HLA-B allotypes, B46 and B73 that both have V76 and N80 motifs, are good ligands for KIR2DL2/3 [47]. In addition to C1 binding, the KIR2DL2/3 can also interact with several C2 allotypes, notably C*0501 and C*0202 [47]. The inhibitory signals triggered by the 2DL2/3-C1 interaction is relatively weaker as compared to those triggered by the 2DL1-C2 interaction [21, 47]. The activating KIR receptor KIR2DS1 has similar Ig-like domains to inhibitory KIR2DL1 and also binds HLA-C2, but with reduced avidity [48]. KIR2DS2 is an activating receptor that has similar Ig-like domains to inhibitory KIR2DL2 but has no detectable avidity for HLA-C1 [21]. KIR2DS4 is the oldest and most prevalent activating receptor and has unique ligand specificity for subsets of HLA-C allotypes carrying C1 or C2 epitopes and HLA-A11 [49].
Since the C1 and C2 are non-overlapping subsets of HLA-C allotypes, individuals can be either C1 homozygous, C2 homozygous or C1/C2 heterozygous. Contrarily, only a subset of HLA-A (HLA-A23, 24, 25 and 32) and HLA-B (40% of the known B allotypes) molecules that carry a Bw4 epitope in a similar region (residues 77-83) function as ligands for the KIR3DL1 receptor [50-52] (Fig. 2). KIR3DL2 binds to only HLA-A3 and A11 allotypes, and the strength of this interaction is highly sensitive to the bound peptide sequence [53, 54]. The KIR2DL4 receptor binds to the extravillous trophoblast-specific HLA-G molecule and induces rapid IFN-γ production that promotes vascularization of the maternal decidua, which provides the placenta with blood and the growing fetus with gases and nutrients [55-58]. In addition to its activation function, the KIR2DL4 receptor carries a single ITIM motif in its cytoplasmic tail and exhibits an inhibitory function [59, 60]. The ligand specificities for KIR2DS2, 2DS3, 2DS5, 3DS1, and 2DL5 remain elusive. Epidemiological studies implicate HLA-Bw4 specificity for KIR3DS1 [61].
Variable KIR and HLA gene families segregate independently, yielding many individuals who express KIR receptors for which they lack HLA class I ligands, and vice versa, thus creating human diversity in the number and type of KIR-HLA gene combinations that can modulate disease outcomes [62] (Fig. 5). Nevertheless, at the population level, there is compelling evidence for coevolution of interacting KIR-HLA pairs [63, 64]. Although most individuals have all four well-defined inhibitory KIR receptors (3DL1, 3DL2, 2DL1 and 2DL2/3), we found only a subset expressing all relevant HLA class I ligands, HLA-Bw4 (3DL1), HLA-A3/11 (3DL2), HLA-C2 (2DL1), and HLA-C1 (2DL2/3) [62]. The majority of Caucasians, Hispanics, and African Americans carry either two or three inhibitory KIR-HLA combinations. Interestingly, one out of five individuals in these populations carries only a single receptor-ligand pair, KIR2DL3+HLA-C1. Environmental invectives, such as viral infections, affecting HLA-C expression in individuals carrying this single KIR-HLA combination can break self-tolerance and may endorse autoimmunity. Importantly, each subject tested in the panel of 759 had at least one of these four inhibitory KIR receptor-HLA class I ligand pairs. This finding is biologically relavent to the the studies showing that the NK cells lacking inhibitory receptors for self-MHC class I molecules were hyporesponsive when compared to the NK cells that had a self-MHC class I ligand [2]. The signals triggered through inhibitory receptors upon binding to the specific self-MHC class I are involved in 'licensing' [2, 65], 'arming' [66], or 'education' [67], a process by which the NK cells acquire functional competence. Therefore, it appears that a minimum of one inhibitory KIR+HLA interaction is crucial for the development of functional NK cells in humans. In summary, interactions of KIR to HLA class I ligands set the threshold of NK cell capacity as well as control the NK cell response.
Viral infection downregulates expression of HLA class I molecules on the surface of infected cells to escape from CTL responses, which renders these class I-negative unhealthy cells as potential targets for NK lysis by the missing-self mechanism (Fig. 1). The impact of NK cells on anti-viral immunity is underscored by the fact that patients with NK cell deficiencies and patients with impaired NK cell activity suffer from recurrent, life-threatening virus infections. Understanding the host KIR-HLA genetic composition that favors successful eradication of virus is especially important for therapeutic vaccination strategies.
Epidemiological studies have identified that host genetic variation at the HLA class I locus has a stronger influence on HIV-1 disease outcome than that of any other genetic locus identified so far [68, 69]. The HLA class I alleles most widely accepted as protective (B*27 and B*57) both carry Bw4, a ligand for KIR3DL1 and possibly for KIR3DS1. About 40% of the observed HLA-B alleles display Bw4, and the remainder HLA-B alleles share the alternative public epitope Bw6. Differential advantage of Bw4-bearing over Bw6-bearing alleles in antigen presentation could neatly connect this broad class I epitopic variation with the increasingly prominent contribution of CTL control and escape in the pathogenesis of AIDS [70, 71]. More recently, a significant association has been described between slower progression in anti-retroviral therapy naive seroconverters and the expression of an activating receptor KIR3DS1, in conjunction with its putative ligand, Bw4-bearing HLA-B molecules with an isoleucine at position 80 (referred to as Bw4I80) in patients of European or African ancestries, which was the first to implicate an activating KIR in NK cell activation that has biological significance in anti-viral immunity [61]. Based on this genetic association, KIR3DS1 on NK cells was proposed to bind to HLA-Bw4I80 on HIV-1 infected target cells, thereby signaling the NK cell to kill the infected target. The KIR3DS1-HLA-Bw4I80 compound genotype was further shown to correlate with lower viral load and protection from opportunistic infections, such as pneumocystis carinii pneumonia, cytomegalovirus (CMV) retinitis, and mycobacterium avium complex [72]. Surprisingly, the inhibitory compound genotype consisting of a subset of KIR3DL1 alleles with high expression and high inhibitory capacity and HLA-Bw4I80 demonstrated significant protection from AIDS progression in the same seroconverted cohorts that showed a protective effect of the KIR3DS1-HLA-Bw4I80 genotype, and it was also associated with lower mean viral loads [61]. Protection by a highly inhibitory KIR-HLA genotype may be attributed to more efficient education of these high-expressing KIR3DL1-positive NK cells during their maturation process, eventually resulting in stronger activation of those NK cells, when their ligand is downregulated. Therefore, the efficient engagement of both activating and inhibitory KIR is likely to be beneficial for the host in viral infection.
Currently 130-170 million people believed to be infected with the hepatitis C virus (HCV), who will either resolve acute infection or progress to chronic infection. The 80% of individuals who become chronically infected have a substantial risk of developing end-stage liver disease, including liver cirrhosis and hepatocellular carcinoma [73]. By comparing the presence or absence of KIR-HLA gene combinations, protection against chronic HCV is shown to be conferred by homozygosity of the genes for the inhibitory receptor KIR2DL3 and its ligand HLA-C with asparagine at position 80 [74, 75]. This protective effect is limited to individuals infected by intravenous drug use or accidental needle-stick injury (low inoculum of virus) but no effect is observed in cases infected by transfusion of blood products, in which the innate immune response is thought to be overwhelmed by the higher infecting inoculum. Since the KIR2DL3 binds HLA-C with a lower avidity than other inhibitory KIR [21], the weak inhibitory signals triggered by this low affinity combination (i.e., the lack of strong NK cell inhibition) presumably being overridden by activating signals of non-variable activating receptors.
NK cells are capable of interacting with a number of cell types through cytokine production and direct cell-cell contact, which have the potential to drive the chronic inflammation. However, accumulating evidence outlined conflicting results of the effect of NK cells in promoting or suppressing inflammatory responses and autoimmune disease development. A protective role for NK cells has been reported in animal models of diabetes [76], colitis [77], and experimental autoimmune encephalomyelitis [78], while in contrast, NK cells have been shown to promote auto-immunity in models of neonatal autoimmune ovarian disease [79] and experimental autoimmune myasthenia gravis [80]. A significant decline in the NK cell population has been reported in the peripheral blood of patients with a variety of autoimmune disorders, but it is unclear if these alterations are a consequence of ongoing inflammation or are an underlying cause of autoimmunity.
Although the cell surface expression and ligands for activating KIR receptors have not been clearly defined, a growing body of genetic epidemiological data reveals the association of distinct activating KIR in the pathogenesis of autoimmune diseases. In these models, the activation signals are proposed to prevail over HLA-dependent inhibition that presumably exacerbates NK cell response [7]. Psoriasis vulgaris is a common inflammatory skin disorder that is strongly associated with KIR2DS1 alone [81, 82] or in combination with HLA-Cw6 [83]. A weak association of the activating receptor-ligand pair KIR2DS2:HLA-CAsn80 is observed in diabetes mellitus [84]. Additionally, the unusual genotype of KIR2DS2 in the absence of its inhibitory counterpart KIR2DL2 is found at high frequency in patients with scleroderma (12%) as compared with controls (2%) [85]. Genotypes encoding a dominant activating KIR genotypes (increased number of activating KIR and less inhibitory KIR-HLA combinations) is found to be associated with certain uveitis conditions including birdshot chorioretinopathy (BCR) [86], Vogt-Koyanagi-Harada (VKH) disease [87], and HLA-B27-associated acute anterior uveitis (AAU) and axial spondyloarthropathy [88].
Elimination of NK cells in mice results in a higher incidence of spontaneous tumors, impaired clearance of inoculated tumor cells, and an increased rate of tumor metastasis [89]. A large body of evidence argues that enhancement of NK cell numbers and function in human cancer patients is associated with increases in tumor clearance and duration of clinical remission [90]. KIR receptors seem to influence malignancies, specifically those associated with viral infections. The presence of KIR3DS1 is shown to be associated with cervical neoplasia progression to cervical cancer, a tumor that is strongly associated with human papilloma viruses (HPV)-16/18 [91]. In contrast, KIR3DS1 and KIR2DS1 genes are shown to protect from developing a severe form of recurrent respiratory papillomatosis (RRP), a rare disease of the larynx and upper airway caused by another HPV strain, HPV-6/11 [92]. The difference in clinical disease expression induced by different HPV strains in 3DS1 carriers could be the interaction of 3DS1 with putative HPV strain-specific ligands leading to either killing of HPV-6/11-infected cells keeping the host from developing RRP or leading to inappropriate tissue-specific hyperresponsiveness promoting growth of cervical cancer, vs. benign respiratory papillomas.
Although NK cells have been known for more than 35 years to have natural ability to kill leukemia and lymphoma cell in rodents [93, 94], their potential therapeutic use in eliminating these hematological cancers in a clinical setting was proposed in 1997 by Valiante and Parham [95]. Hematopoietic stem cells (HSC) transplantation has been increasingly adopted as a therapy for a number of malignant and nonmalignant hematologic diseases, including leukemia, lymphoma, aplastic anemia, thalassemia major, immunodeficiency diseases, and severe combined immunodeficiency (SCID) [96-98]. Following allogeneic HSCT, the progeny of the donor stem cells repopulate the entire hematopoietic system of the recipient. The T cells of donor origin are critical for promoting engraftment and eradicating malignant cells, but may cause an adverse reaction such as graft rejection and graft-versus-host disease (GVHD), which is an attack on recipient tissues. To minimize T cell alloreactivity, HSCT is performed with HLA-matched donors. An HLA-matched donor can be KIR mismatched, since the KIR gene family is not physically linked to HLA gene family. NK cells are the first lymphocyte population to appear in peripheral blood shortly after HSCT, and the KIR receptor repertoires of reconstituted NK cells are consistently of donor type [99].
The donor-derived NK cells can be alloreactive, if their inhibitory KIRs do not see a relevant HLA class I ligand in the recipient that was present in the donor. Such alloreactive NK cells greatly contribute many potential benefits, including i) decreased rates of GVHD, ii) decreased rates of graft rejection mediated by NK lysis of host T cells, iii) decreased relapse, iv) improved engraftment mediated by NK-cell release of hematopoietic cytokines, and v) enhanced immune reconstitution and decreased infectious complications mediated by NK-cell antiviral activity. Such beneficial NK cell alloreactivity, which can be predicted from the differences in KIR-binding HLA class I ligands between donor and recipient based on their HLA class I type, was first described for HLA haploidentical transplantation by the use of an extensively T cell-depleted graft in acute myeloid leukemia (AML) patients [100] and later investigated in other transplantation settings [101, 102]. Patients with myeloid malignancies are more responsive to treatment than those with lymphoid malignancies. Several studies have suggested that adult acute lymphoid leukemia (ALL) is not as susceptible to KIR ligand mismatched haploidentical allogeneic HSCT [100, 103-105]. However, alloreactive NK cell-mediated effects can impact childhood ALL [106]. Beneficial effects from KIR-ligand mismatch have not been seen in the T cell-replete setting [107, 108].
In addition to the recipient lacking HLA class I ligands for the donor-derived NK cells, expression of activating KIR receptors on donor NK cells is also shown to influence HSCT outcome. Compared to donors with AA genotypes (express one or no activating KIR), the Bx genotype (express 1-6 activating KIRs) donors are shown to contribute significantly superior relapse protection and improved disease-free survival for AML patients [109]. Gene content motif analyses further reveal that the centromeric and telomeric B haplotype-specific motifs both contribute to relapse protection and improved survival, but centromeric B homozygosity (Cen-B/B) has the strongest independent effect [109]. Further studies are required to determine if the clinical benefit conferred by Cen-B/B is caused by a single KIR gene (such as 2DS2, 2DS3 or KIR2DL2) or by the combination of specific KIR genes.
Recent studies reveal the strongest clinical impact of telomeric B haplotype-specific activating KIR genes [110, 111]. Compared with KIR3DS1-negative donors, a donor with KIR3DS1 is shown to be associated with lower-grade II-IV acute GVHD, but not with relapse [110]. Furthermore, grade II-IV acute GVHD, overall mortality, and transplantation-related mortality all decreased as the number of copies of donor KIR3DS1 increased, with the lowest failure rate occurring among patients homozygous for donor KIR3DS1 [110]. Functional experiments reveal that the activating KIR2DS1 plays a substantial role in mediating alloreactivity and confers an advantage in the ability of NK cell alloreactivity to kill dendritic cells and T cell blasts [111]. In summary, knowing the KIR genotype of the donor, and HLA types of both the donor and recipient, it is possible to predict the degree of KIR-HLA interactions that may determine an enhanced ability to limit GVHD and improve engraftment for certain leukemias.
Emerging understanding of KIR and MHC loci provides new insights into their diversity between individuals, human populations, and distinct species. NK cells are more than simple innate killers and participate in adoptive immunity to infections, cancer and transplantation. Interactions between the products of the independently segregating polymorphic KIR and HLA loci regulate NK cell effector function in distinguishing unhealthy cell targets form the healthy host cells. Although certain KIR-HLA compound genotypes have been implicated in human disease, it is important to develop studies to understand the impact of KIR and HLA sequence variants, and their combinations on receptor-ligand expression, recognition, development, signaling, effector function, and impact on disease susceptibility and resistance. Such sequence diversity studies must also be extended to different ethnic populations to understand the natural selection of KIR-HLA compound genotypes that might favor population adaptation and survival. Further studies on the functional characterization of the NK subsets expressing distinct KIR receptor repertoire at healthy and disease state will be of great interest and may provide a better understanding of the NK tolerance and pathogenesis of disease. Functional studies aiming to investigate the impact of variable KIR-HLA interactions on NK cell development, differentiation, education, homeostasis, longevity, and functional heterogeneity are essential to underpin their potential for rational use in humans and optimizing the use of NK cells for treatment of disease.
References
1. Narni-Mancinelli E, Vivier E, Kerdiles YM. The 'T-cell-ness' of NK cells: unexpected similarities between NK cells and T cells. Int Immunol. 2011; 23:427–431. PMID: 21665959.

2. Kim S, Poursine-Laurent J, Truscott SM, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005; 436:709–713. PMID: 16079848.

3. Sun JC, Beilke JN, Lanier LL. Immune memory redefined: characterizing the longevity of natural killer cells. Immunol Rev. 2010; 236:83–94. PMID: 20636810.

4. Bryceson YT, Long EO. Line of attack: NK cell specificity and integration of signals. Curr Opin Immunol. 2008; 20:344–352. PMID: 18439809.

5. Lanier LL. Natural killer cell receptor signaling. Curr Opin Immunol. 2003; 15:308–314. PMID: 12787756.

6. Valiante NM, Uhrberg M, Shilling HG, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997; 7:739–751. PMID: 9430220.

8. McQueen KL, Parham P. Variable receptors controlling activation and inhibition of NK cells. Curr Opin Immunol. 2002; 14:615–621. PMID: 12183162.

9. Vivier E, Raulet DH, Moretta A, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011; 331:44–49. PMID: 21212348.

10. Raulet DH, Vance RE, McMahon CW. Regulation of the natural killer cell receptor repertoire. Annu Rev Immunol. 2001; 19:291–330. PMID: 11244039.

11. Ljunggren HG, Karre K. In search of the 'missing self': MHC molecules and NK cell recognition. Immunol Today. 1990; 11:237–244. PMID: 2201309.

12. Abi-Rached L, Moesta AK, Rajalingam R, Guethlein LA, Parham P. Human-specific evolution and adaptation led to major qualitative differences in the variable receptors of human and chimpanzee natural killer cells. PLoS Genet. 2010; 6:e1001192. PMID: 21079681.

13. Wilson MJ, Torkar M, Haude A, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000; 97:4778–4783. PMID: 10781084.

14. Uhrberg M, Parham P, Wernet P. Definition of gene content for nine common group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes. Immunogenetics. 2002; 54:221–229. PMID: 12136333.

15. Hsu KC, Liu XR, Selvakumar A, Mickelson E, O'Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. 2002; 169:5118–5129. PMID: 12391228.

16. Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006; 203:633–645. PMID: 16533882.

17. Whang DH, Park H, Yoon JA, Park MH. Haplotype analysis of killer cell immunoglobulin-like receptor genes in 77 Korean families. Hum Immunol. 2005; 66:146–154. PMID: 15695000.

18. Middleton D, Meenagh A, Gourraud PA. KIR haplotype content at the allele level in 77 Northern Irish families. Immunogenetics. 2007; 59:145–158. PMID: 17200871.

19. Shilling HG, Guethlein LA, Cheng NW, et al. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002; 168:2307–2315. PMID: 11859120.

20. Garcia CA, Robinson J, Guethlein LA, Parham P, Madrigal JA, Marsh SG. Human KIR sequences 2003. Immunogenetics. 2003; 55:227–239. PMID: 12838379.

21. Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998; 161:571–577. PMID: 9670929.
22. Gardiner CM, Guethlein LA, Shilling HG, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001; 166:2992–3001. PMID: 11207248.

23. Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005; 175:5222–5229. PMID: 16210627.

24. Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Variable NK cell receptors exemplified by human KIR3DL1/S1. J Immunol. 2011; 187:11–19. PMID: 21690332.

25. Norman PJ, Abi-Rached L, Gendzekhadze K, et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007; 39:1092–1099. PMID: 17694054.

26. Vivian JP, Duncan RC, Berry R, et al. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature. 2011; 479:401–405. PMID: 22020283.

27. Hsu KC, Chida S, Geraghty DE, Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev. 2002; 190:40–52. PMID: 12493005.

28. Yawata M, Yawata N, Abi-Rached L, Parham P. Variation within the human killer cell immunoglobulin-like receptor (KIR) gene family. Crit Rev Immunol. 2002; 22:463–482. PMID: 12803322.
29. Norman PJ, Abi-Rached L, Gendzekhadze K, et al. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. 2009; 19:757–769. PMID: 19411600.

30. Ordonez D, Meenagh A, Gómez-Lozano N, Castanño J, Middleton D, Vilches C. Duplication, mutation and recombination of the human orphan gene KIR2DS3 contribute to the diversity of KIR haplotypes. genes Immun. 2008; 9:431–437. PMID: 18480828.

31. Ashouri E, Farjadian S, Reed EF, Ghaderi A, Rajalingam R. KIR gene content diversity in four Iranian populations. Immunogenetics. 2009; 61:483–492. PMID: 19521696.

32. Uhrberg M, Valiante NM, Shum BP, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997; 7:753–763. PMID: 9430221.

33. Jiang K, Zhu FM, Lv QF, Yan LX. Distribution of killer cell immunoglobulin-like receptor genes in the Chinese Han population. Tissue Antigens. 2005; 65:556–563. PMID: 15896204.

34. Yawata M, Yawata N, McQueen KL, et al. Predominance of group A KIR haplotypes in Japanese associated with diverse NK cell repertoires of KIR expression. Immunogenetics. 2002; 54:543–550. PMID: 12439616.

35. Gendzekhadze K, Norman PJ, Abi-Rached L, Layrisse Z, Parham P. High KIR diversity in Amerindians is maintained using few gene-content haplotypes. Immunogenetics. 2006; 58:474–480. PMID: 16738943.

36. Ewerton PD, Leite Mde M, Magalhães M, Sena L, Melo dos Santos EJ. Amazonian Amerindians exhibit high variability of KIR profiles. Immunogenetics. 2007; 59:625–630. PMID: 17551723.

37. Toneva M, Lepage V, Lafay G, et al. Genomic diversity of natural killer cell receptor genes in three populations. Tissue Antigens. 2001; 57:358–362. PMID: 11380947.

38. Rajalingam R, Du Z, Meenagh A, et al. Distinct diversity of KIR genes in three southern Indian populations: comparison with world populations revealed a link between KIR gene content and pre-historic human migrations. Immunogenetics. 2008; 60:207–217. PMID: 18369612.

39. Rajalingam R, Krausa P, Shilling HG, et al. Distinctive KIR and HLA diversity in a panel of north Indian Hindus. Immunogenetics. 2002; 53:1009–1019. PMID: 11904677.

40. Kulkarni S, Single RM, Martin MP, et al. Comparison of the rapidly evolving KIR locus in Parsis and natives of India. Immunogenetics. 2008; 60:121–129. PMID: 18351333.

41. Marsh SG, Albert ED, Bodmer WF, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010; 75:291–455. PMID: 20356336.
42. Klein J, Sato A. The HLA system. Second of two parts. N Engl J Med. 2000; 343:782–786. PMID: 10984567.

43. Klein J, Sato A. The HLA system. First of two parts. N Engl J Med. 2000; 343:702–709. PMID: 10974135.
44. Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci U S A. 1993; 90:12000–12004. PMID: 8265660.

45. Wagtmann N, Biassoni R, Cantoni C, et al. Molecular clones of the p58 NK cell receptor reveal immunoglobulin-related molecules with diversity in both the extra- and intracellular domains. Immunity. 1995; 2:439–449. PMID: 7749980.

46. Winter CC, Long EO. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol. 1997; 158:4026–4028. PMID: 9126959.
47. Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008; 180:3969–3979. PMID: 18322206.

48. Stewart CA, Laugier-Anfossi F, Vély F, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A. 2005; 102:13224–13229. PMID: 16141329.

49. Graef T, Moesta AK, Norman PJ, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009; 206:2557–2572. PMID: 19858347.

50. Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995; 181:1133–1144. PMID: 7532677.

51. Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994; 180:1235–1242. PMID: 7931060.

52. Thananchai H, Gillespie G, Martin MP, et al. Cutting edge: allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007; 178:33–37. PMID: 17182537.

53. Pende D, Biassoni R, Cantoni C, et al. The natural killer cell receptor specific for HLA-A allotypes: a novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulphide-linked dimer. J Exp Med. 1996; 184:505–518. PMID: 8760804.

54. Hansasuta P, Dong T, Thananchai H, et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004; 34:1673–1679. PMID: 15162437.

55. Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med. 1999; 189:1093–1100. PMID: 10190900.

56. Moffett-King A. Natural killer cells and pregnancy. Nat Rev Immunol. 2002; 2:656–663. PMID: 12209134.

57. Rajagopalan S, Bryceson YT, Kuppusamy SP, et al. Activation of NK cells by an endocytosed receptor for soluble HLA-G. PLoS Biol. 2006; 4:e9. PMID: 16366734.

58. Kikuchi-Maki A, Yusa S, Catina TL, Campbell KS. KIR2DL4 is an IL-2-regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFN-gamma production. J Immunol. 2003; 171:3415–3425. PMID: 14500636.
59. Faure M, Long EO. KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory potential. J Immunol. 2002; 168:6208–6214. PMID: 12055234.

60. Ponte M, Cantoni C, Biassoni R, et al. Inhibitory receptors sensing HLA-G1 molecules in pregnancy: decidua-associated natural killer cells express LIR-1 and CD94/NKG2A and acquire p49, an HLA-G1-specific receptor. Proc Natl Acad Sci U S A. 1999; 96:5674–5679. PMID: 10318943.

61. Martin MP, Gao X, Lee JH, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002; 31:429–434. PMID: 12134147.

62. Du Z, Gjertson DW, Reed EF, Rajalingam R. Receptor-ligand analyses define minimal killer cell Ig-like receptor (KIR) in humans. Immunogenetics. 2007; 59:1–15. PMID: 17103212.

63. Single RM, Martin MP, Gao X, et al. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet. 2007; 39:1114–1119. PMID: 17694058.

64. Gendzekhadze K, Norman PJ, Abi-Rached L, et al. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci U S A. 2009; 106:18692–18697. PMID: 19837691.

65. Yokoyama WM, Kim S. How do natural killer cells find self to achieve tolerance? Immunity. 2006; 24:249–257. PMID: 16546094.

66. Raulet DH, Vance RE. Self-tolerance of natural killer cells. Nat Rev Immunol. 2006; 6:520–531. PMID: 16799471.

67. Anfossi N, Andre P, Guia S, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006; 25:331–342. PMID: 16901727.

68. Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003; 54:535–551. PMID: 12525683.

69. Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007; 317:944–947. PMID: 17641165.

70. Goulder PJ, Brander C, Tang Y, et al. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 2001; 412:334–338. PMID: 11460164.

71. Betts MR, Nason MC, West SM, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006; 107:4781–4789. PMID: 16467198.

72. Qi Y, Martin MP, Gao X, et al. KIR/HLA pleiotropism: protection against both HIV and opportunistic infections. PLoS Pathog. 2006; 2:e79. PMID: 16933987.

73. Alter HJ, Seeff LB. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin Liver Dis. 2000; 20:17–35. PMID: 10895429.

74. Khakoo SI, Thio CL, Martin MP, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004; 305:872–874. PMID: 15297676.

75. Romero V, Azocar J, Zúñiga J, et al. Interaction of NK inhibitory receptor genes with HLA-C and MHC class II alleles in Hepatitis C virus infection outcome. Mol Immunol. 2008; 45:2429–2436. PMID: 18289678.

76. Lee IF, Qin H, Trudeau J, Dutz J, Tan R. Regulation of autoimmune diabetes by complete Freund's adjuvant is mediated by NK cells. J Immunol. 2004; 172:937–942. PMID: 14707066.

77. Fort MM, Leach MW, Rennick DM. A role for NK cells as regulators of CD4+ T cells in a transfer model of colitis. J Immunol. 1998; 161:3256–3261. PMID: 9759840.
78. Xu W, Fazekas G, Hara H, Tabira T. Mechanism of natural killer (NK) cell regulatory role in experimental autoimmune encephalomyelitis. J Neuroimmunol. 2005; 163:24–30. PMID: 15885305.

79. Setiady YY, Pramoonjago P, Tung KS. Requirements of NK cells and proinflammatory cytokines in T cell-dependent neonatal autoimmune ovarian disease triggered by immune complex. J Immunol. 2004; 173:1051–1058. PMID: 15240693.

80. Shi FD, Wang HB, Li H, et al. Natural killer cells determine the outcome of B cell-mediated autoimmunity. Nat Immunol. 2000; 1:245–251. PMID: 10973283.

81. Suzuki Y, Hamamoto Y, Ogasawara Y, et al. Genetic polymorphisms of killer cell immunoglobulin-like receptors are associated with susceptibility to psoriasis vulgaris. J Invest Dermatol. 2004; 122:1133–1136. PMID: 15140215.

82. Holm SJ, Sakuraba K, Mallbris L, Wolk K, Stahle M, Sanchez FO. Distinct HLA-C/KIR genotype profile associates with guttate psoriasis. J Invest Dermatol. 2005; 125:721–730. PMID: 16185272.

83. Luszczek W, Mańczak M, Cisło M, et al. Gene for the activating natural killer cell receptor, KIR2DS1, is associated with susceptibility to psoriasis vulgaris. Hum Immunol. 2004; 65:758–766. PMID: 15310528.
84. van der Slik AR, Koeleman BP, Verduijn W, Bruining GJ, Roep BO, Giphart MJ. KIR in type 1 diabetes: disparate distribution of activating and inhibitory natural killer cell receptors in patients versus HLA-matched control subjects. Diabetes. 2003; 52:2639–2642. PMID: 14514651.
85. Momot T, Koch S, Hunzelmann N, et al. Association of killer cell immunoglobulin-like receptors with scleroderma. Arthritis Rheum. 2004; 50:1561–1565. PMID: 15146426.

86. Levinson RD, Du Z, Luo L, et al. Combination of KIR and HLA gene variants augments the risk of developing birdshot chorioretinopathy in HLA-A*29-positive individuals. Genes Immun. 2008; 9:249–258. PMID: 18340360.

87. Levinson RD, Okada AA, Ashouri E, Keino H, Rajalingam R. Killer cell immunoglobulin-like receptor gene-cluster 3DS1-2DL5-2DS1-2DS5 predisposes susceptibility to Vogt-Koyanagi-Harada syndrome in Japanese individuals. Hum Immunol. 2010; 71:192–194. PMID: 19897003.

88. Levinson RD, Martin TM, Luo L, et al. Killer Cell Immunoglobulin-like receptors in HLA-B27-associated acute anterior uveitis, with and without axial spondyloarthropathy. Invest Ophthalmol Vis Sci. 2010; 51:1505–1510. PMID: 19850842.

89. Waldhauer I, Steinle A. NK cells and cancer immunosurveillance. Oncogene. 2008; 27:5932–5943. PMID: 18836474.

90. Zamai L, Ponti C, Mirandola P, et al. NK cells and cancer. J Immunol. 2007; 178:4011–4016. PMID: 17371953.

91. Carrington M, Wang S, Martin MP, et al. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J Exp Med. 2005; 201:1069–1075. PMID: 15809352.

92. Bonagura VR, Du Z, Ashouri E, et al. Activating killer cell immunoglobulin-like receptors 3DS1 and 2DS1 protect against developing the severe form of recurrent respiratory papillomatosis. Hum Immunol. 2010; 71:212–219. PMID: 19861144.

93. Kiessling R, Klein E, Pross H, Wigzell H. "Natural" killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur J Immunol. 1975; 5:117–121. PMID: 1086218.

94. Herberman RB, Nunn ME, Holden HT, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. II. Characterization of effector cells. Int J Cancer. 1975; 16:230–239. PMID: 1080480.

95. Valiante NM, Parham P. Natural killer cells, HLA class I molecules, and marrow transplantation. Biol Blood Marrow Transplant. 1997; 3:229–235. PMID: 9450917.
96. Appelbaum FR. The current status of hematopoietic cell transplantation. Annu Rev Med. 2003; 54:491–512. PMID: 12414918.

97. Thomas ED. History, current results, and research in marrow transplantation. Perspect Biol Med. 1995; 38:230–237. PMID: 7899057.

99. Shilling HG, Young N, Guethlein LA, et al. Genetic control of human NK cell repertoire. J Immunol. 2002; 169:239–247. PMID: 12077250.

100. Ruggeri L, Capanni M, Urbani E, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002; 295:2097–2100. PMID: 11896281.

101. Giebel S, Locatelli F, Lamparelli T, et al. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood. 2003; 102:814–819. PMID: 12689936.

102. Davies SM, Ruggieri L, DeFor T, et al. Evaluation of KIR ligand incompatibility in mismatched unrelated donor hematopoietic transplants. Killer immunoglobulin-like receptor. Blood. 2002; 100:3825–3827. PMID: 12393440.
103. Kawase T, Matsuo K, Kashiwase K, et al. HLA mismatch combinations associated with decreased risk of relapse: implications for the molecular mechanism. Blood. 2009; 113:2851–2858. PMID: 18997170.

104. Clausen J, Wolf D, Petzer AL, et al. Impact of natural killer cell dose and donor killer-cell immunoglobulin-like receptor (KIR) genotype on outcome following human leucocyte antigen-identical haematopoietic stem cell transplantation. Clin Exp Immunol. 2007; 148:520–528. PMID: 17493020.

105. Willemze R, Rodrigues CA, Labopin M, et al. KIR-ligand incompatibility in the graft-versus-host direction improves outcomes after umbilical cord blood transplantation for acute leukemia. Leukemia. 2009; 23:492–500. PMID: 19151783.

106. Pende D, Marcenaro S, Falco M, et al. Anti-leukemia activity of alloreactive NK cells in KIR ligand-mismatched haploidentical HSCT for pediatric patients: evaluation of the functional role of activating KIR and redefinition of inhibitory KIR specificity. Blood. 2009; 113:3119–3129. PMID: 18945967.

107. Lowe EJ, Turner V, Handgretinger R, et al. T-cell alloreactivity dominates natural killer cell alloreactivity in minimally T-cell-depleted HLA-non-identical paediatric bone marrow transplantation. Br J Haematol. 2003; 123:323–326. PMID: 14531915.

108. Farag SS, Bacigalupo A, Eapen M, et al. The effect of KIR ligand incompatibility on the outcome of unrelated donor transplantation: a report from the center for international blood and marrow transplant research, the European blood and marrow transplant registry, and the Dutch registry. Biol Blood Marrow Transplant. 2006; 12:876–884. PMID: 16864058.

109. Cooley S, Weisdorf DJ, Guethlein LA, et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood. 2010; 116:2411–2419. PMID: 20581313.

110. Venstrom JM, Gooley TA, Spellman S, et al. Donor activating KIR3DS1 is associated with decreased acute GVHD in unrelated allogeneic hematopoietic stem cell transplantation. Blood. 2010; 115:3162–3165. PMID: 20124216.

111. Sivori S, Carlomagno S, Falco M, Romeo E, Moretta L, Moretta A. Natural killer cells expressing the KIR2DS1-activating receptor efficiently kill T-cell blasts and dendritic cells: implications in haploidentical HSCT. Blood. 2011; 117:4284–4292. PMID: 21355085.

112. Ashouri E, Farjadian S, Reed EF, Ghaderi A, Rajalingam R. KIR gene content diversity in four Iranian populations. Immunogenetics. 2009; 61:483–492. PMID: 19521696.

Fig. 1
Professional killer cell response. Natural killer (NK) cells and cytotoxic T cells (CTL) are professional killer cells and share several common features but differ by their HLA class I-specific receptors that are used to distinguish unhealthy targets from the healthy host cells. CTLs express activating T cell receptors (TCR) while NK cells express both inhibitory and activating receptors. No activation is triggered if the TCR recognize a self-peptide laden HLA class I molecule (a). TCR can trigger cytolysis if it detects a viral peptide loaded in the groove of HLA class I molecules of infected cells (b). Contrarily, the inhibitory receptors of NK cell recognize HLA class I molecules and trigger signals that stop spontaneous lytic activity of NK cells (c). By expressing normal levels of HLA class I molecules, the healthy cells are tolerant to NK cell lysis. Downregulation of HLA class I expression due to tumor transformation or viral infection relieves the inhibitory influence on NK cells, permitting NK cells to lyse the unhealthy target cells (d). The NK cell lysis can be augmented by further interactions between the activating receptors and putative ligands expressed upon infection or transformation (e).
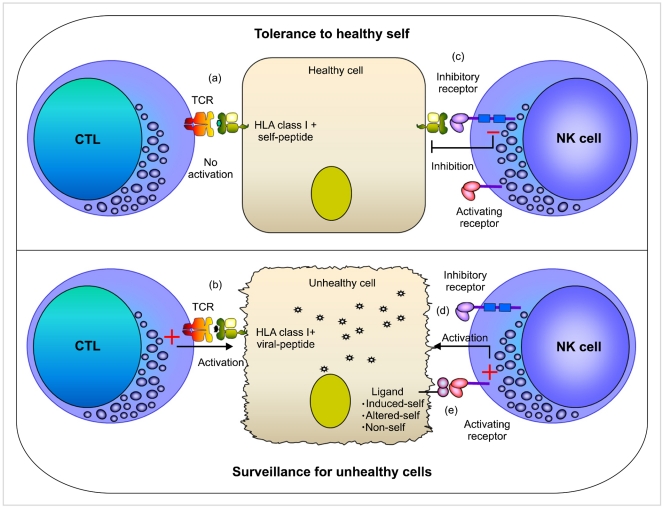
Fig. 2
Killer cell Immunoglobulin-like Receptors (KIR). Fourteen distinct KIR receptors have been characterized in humans that comprise either two (2D) or three (3D) extracellular immunoglobulin-like domains and either a long (L) or short (S) cytoplasmic tail. Six KIR receptors are activating types and the remaining KIR are inhibitory types. The ITIM motifs in the cytoplasmic tails of inhibitory KIRs are shown as blue boxes, and positively charged residues in the transmembrane regions of activating KIRs are shown as yellow circles. The inhibitory KIR receptors bind to distinct HLA class I allotypes and the ligands for most activating KIR receptors are unknown. The number of protein sequence variants characterized to date for each KIR receptor is provided. This data was extracted from the IPD-KIR database (http://www.ebi.ac.uk/ipd/kir/stats.html; Release 2.4.0; April 2011) that provides a centralized repository for human KIR sequences.
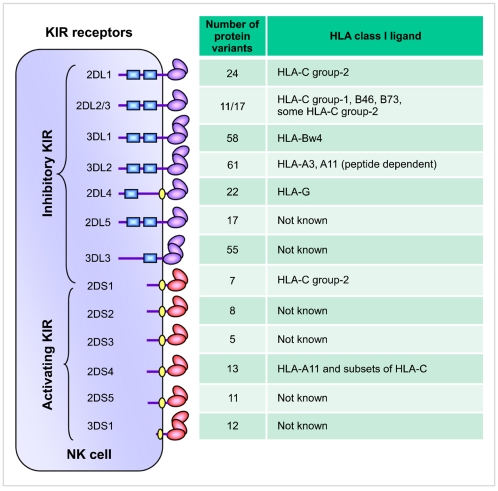
Fig. 3
KIR haplotypes differ by gene content. Map of KIR haplotypes as determined by sequencing genomic clones and haplotype segregation analysis in families. Haplotype 1 represents group-A KIR haplotype and the remainder group-B haplotypes (haplotypes 2-22). The framework genes, present in all haplotypes are shown in dark boxes; genes encoding activating KIR are in orange boxes; and those for inhibitory receptors are in blue boxes. The KIR2DP1 and 3DP1 are pseudogenes that do not express a receptor. Maps are not drawn to scale.
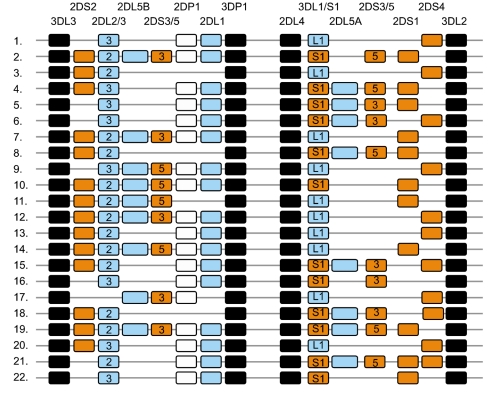
Fig. 4
Centromeric and telomeric halves of KIR haplotypes. A stretch of 14 kb DNA with numerous L1 repeats that interconnects KIR3DP1 and KIR2DL4 divides the KIR haplotype into two halves. The centromeric half is delimited by 3DL3 and 3DP1, while the telomeric half is delimited by 2DL4 and 3DL2. Multiple reciprocal meiotic recombination events between 3DP1 and 2DL4 shuffle the centromeric (c) and telomeric (t) motifs, and thus diversify gene content of KIR haplotypes across individuals and populations. Most of the KIR gene content haplotypes published to date can be explained by the recombination of these 10 centromeric and 10 telomeric gene content motifs. The framework genes, present in all haplotypes are shown in dark boxes; genes encoding activating KIR are in orange boxes; and those for inhibitory receptors are in blue boxes. The KIR2DP1 and 3DP1 are pseudogenes that do not express a receptor. Letter 'A' in gene-content motif identification indicates parts of group-A haplotypes, while 'B' indicates parts of group-B haplotypes.
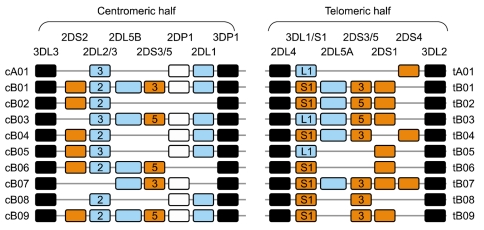
Fig. 5
KIR and HLA gene families are unlinked and located on different human chromosomes, 19 and 6 respectively. Both gene families evolve rapidly and feature substantial variation between haplotypes in the number and type of genes. Panel I illustrates variation between parents in KIR and HLA haplotypes. Panel II illustrates all four possible types of gametes from each parent assorted with different combination of one KIR haplotype and one HLA haplotype. Random associations of gametes form zygotes of the next generation carrying one of the 16 possible combinations of paternal and maternal KIR and HLA haplotypes, producing substantial diversity between offspring in the number and type of inhibitory KIR-HLA combinations and activating KIR genes inherited (Panel III).
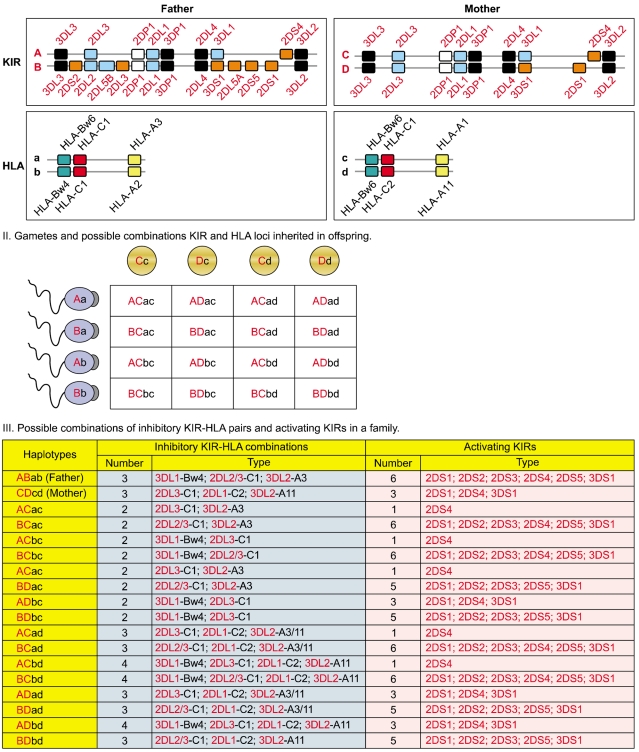
Fig. 6
A and B haplotype distribution varies across distinct populations. The A and B KIR haplotypes are approximately equally distributed in Caucasians and Africans. In contrast, the prevalence of A haplotypes dominates over the B haplotypes in Northeast Asians (Chinese, Japanese, and Koreans) and vice versa in the natives of America, Australia, and India. As we described elsewhere, the frequency of group A and B KIR haplotypes were predicted from the KIR gene content data published previously for other populations [112].




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download