INTRODUCTION
Karyotypic investigations, including fluorescence
in situ hybridization (FISH), have become increasingly important in the detection of hematologic malignancies [
1,
2]. In cases where cytogenetic analysis is hampered by low
in vitro mitotic activity of cancer cells, poor chromosome morphology, considerable complexity, or a normal karyotype, FISH analysis has provided a rapid and reliable detection of specific abnormalities in both mitotic and interphase cells [
1,
2]. Both conventional G-banded karyotype and FISH analyses are currently integral components in the management of patients with hematologic malignancies.
However, in view of limited laboratory and health care resources, FISH can be a labor-intensive, time-consuming, and expensive procedure, particularly if a specific abnormality has already been detected by G-banded karyotype. Therefore, the FISH approach should be strategically planned in order to contribute information additional to that provided by conventional G-banded karyotype.
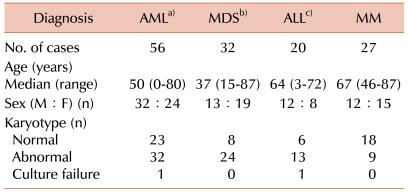
The aim of this study was to evaluate the clinical utility of FISH in addition to G-banded karyotype and to propose a practical approach for FISH in the detection of hematologic malignancies, including acute myeloid leukemia (AML), myelodysplastic syndrome (MDS), acute lymphoblastic leukemia (ALL), and multiple myeloma (MM).
Go to :

RESULTS
FISH studies confirmed the corresponding abnormalities identified by G-banded karyotype in all of the AML samples. Additional abnormalities were detected by FISH in only two cases (4%). In 1 case with unsuccessful culture, AML1 (RUNX1) gain was identified in 66% of interphase cells by FISH. In the other case, characterized by a complex karyotype with marker chromosomes and abnormalities of chromosome 5, 17, 18, and 19, FISH detected MLL gain in 80% of interphase cells.
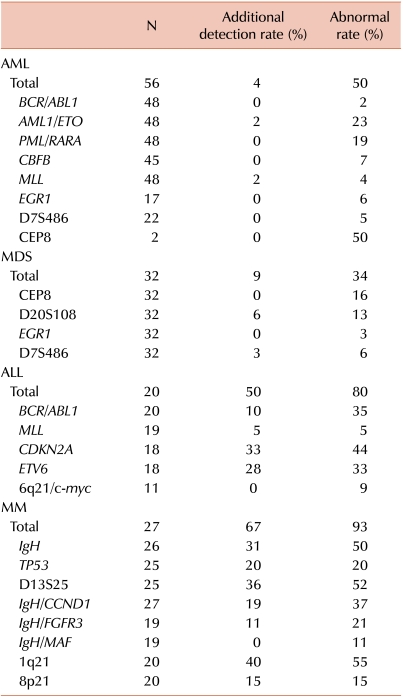
Table 2
Abnormalities detected by FISH in addition to G-banded karyotype.

FISH confirmed the results of both normal and abnormal karyotype identified by G-banded karyotype. Abnormalities additional to those identified by G-banded karyotype were identified by FISH in 3 patients with normal karyotype (2 patients, 20q deletion in 5-6% of interphase cells; 1 patient, 7q deletion in 3% of interphase cells).
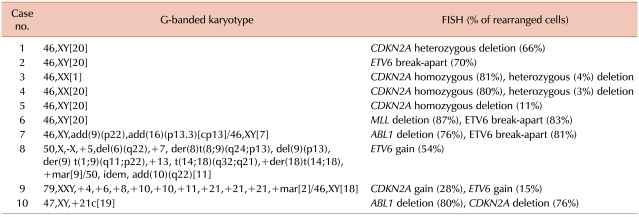
Clonal abnormalities were found in 65% and 80% of patients by G-banded karyotype and FISH, respectively. Additional abnormalities were identified by FISH in 50% of patients. Compared with G-banded karyotype, CDKN2A and ETV6 FISH revealed additional genetic aberrations in 33% and 28% of cases, respectively. Two patients showed ETV6 gain using FISH, corresponding to the hyperdiploidy or hypertriploidy by G-banded karyotype. In 2 patients, ABL1 deletions unassociated with t(9;22) were identified by FISH.
Table 3
Additional genetic aberrations identified by FISH in ALL.

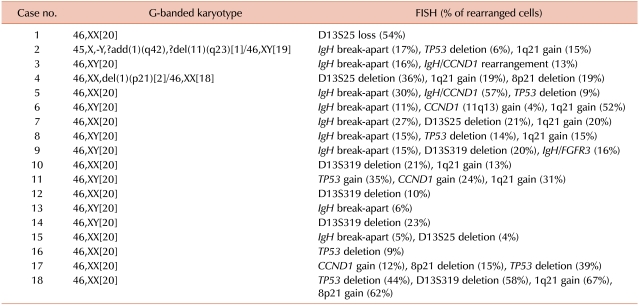
Clonal abnormalities were detected in 33% and 93% of patients by G-banded karyotype and FISH, respectively. Additional abnormalities were identified by FISH in 67% of patients, and among these, 89% had a normal karyotype as determined by G-banded karyotype. FISH was of benefit in detecting IgH, D13S25, TP53, and 1q21 rearrangements that were not detected by G-banded karyotype (31%, 36%, 20%, and 40%, respectively).
Table 4
Additional genetic aberrations identified by FISH in MM.

Go to :

DISCUSSION
In this study, the percentages of additional genetic aberrations identified by FISH (which were not detected by G-banded karyotype) were 4%, 9%, 50%, and 67% in AML, MDS, ALL, and MM, respectively.
In AML and MDS, FISH did not add any relevant information to that already provided by G-banded karyotype with regard to specific chromosomal abnormalities. Similar to our result, a previous study showed that AML FISH profile tests revealed additional genetic abnormalities only in 8% of cases [
4]. Another study showed that the discrepancy between G-banded karyotype and FISH for the diagnosis of AML is 7% [
5]. Previous studies evaluating the utility of FISH in cases of MDS reported the detection of 6% or fewer abnormalities by FISH in addition to those detected by G-banded karyotype [
6-
13]. Other studies, however, have detected up to approximately 15% of additional abnormalities using FISH [
14-
16]. One study suggested that FISH testing may be informative only in MDS cases with culture failure or intermediate- to high-grade MDS cases with normal karyotype, indicating that cases with low-grade MDS and normal karyotypes do not appear to benefit from FISH testing [
13]. Taken together, in the setting of adequate cytogenetic study, the sensitivity of the two techniques in detecting clinically significant chromosomal abnormalities seems to be similar, and FISH may have limited utility in the cases of AML or MDS.
In contrast, FISH, particularly in the cases of lymphoid malignancies, becomes an invaluable tool for identifying specific genetic changes other than G-banded karyotype. One study showed that ALL FISH profile tests revealed additional genetic aberrations not detected by G-banded karyotype in up to 49% of cases, and among these,
ETV6/RUNX1 (
TEL/AML1) abnormalities were frequently detected (44%), followed by the abnormal
CDKN2A (25%) and hyperdiploidy (18%) [
4]. A further study also demonstrated that G-banded karyotype failed to detect a considerable part of the
ETV6/RUNX1 (
TEL/AML1) translocation (sensitivity 6%) [
17]. As expected, FISH is of benefit in the detection of
ETV6 and
CDKN2A rearrangements, since these rearrangements are cytogenetically cryptic and invisible. On the other hand, the sensitivity of G-banded karyotype for the detection of
BCR/ABL1 and
MLL rearrangements in ALL is relatively high (80% and 85%, respectively) [
17]. Consistent with previous studies, our study showed that in ALL,
ETV6 and
CDKN2A genetic abnormalities, which were not identified by G-banded karyotype, could be additionally detected by FISH (28% and 33%, respectively).
As reported previously, FISH is useful for improving the detection rate of genetic abnormalities in MM, whereas conventional cytogenetics detects only 30-50% of abnormalities due to the low
in vitro mitotic index of abnormal clones [
18-
25]. In this study, the detection rate of genomic aberrations in MM increased from 67% to 93% using FISH, compared with G-banded karyotype.
FISH has been performed extensively to detect genomic aberrations in hematologic malignancies; however, in view of limited laboratory resources, it may be an expensive procedure. Here, we propose a strategy for cost-effective FISH utilization based on our results (
Fig. 1). Importantly, this strategy should be based on the premise of cytogenetic adequacy (analyzing more than 20 consecutive, well-stained, well-spread metaphases). If a sufficient number of metaphases with morphology good enough to detect microscopic abnormalities cannot be analyzed, this strategy should not be applied.
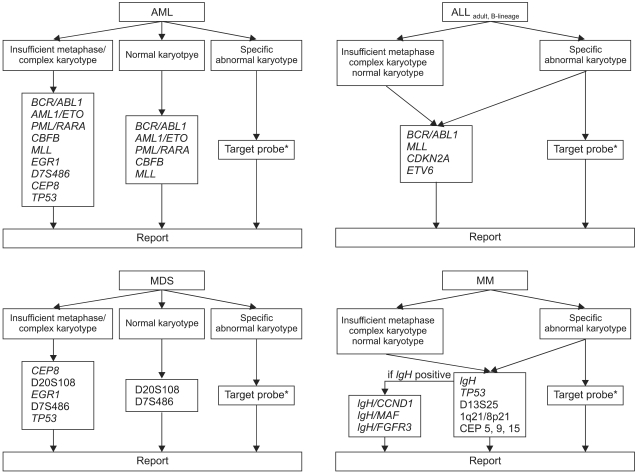 | Fig. 1Proposal for a cost-effective utilization of FISH in hematologic malignancies (*corresponded to specific chromosomal abnormalities identified by G-banded karyotype). 
|
With respect to ALL and MM, routine FISH analysis is needed, irrespective of karyotypic results (normal or abnormal), since, as shown in this study, FISH can provide relevant information additional to that provided by G-banded karyotype (
Tables 2-
4). In order to detect aneuploidy, it may be necessary to include centromere probes for chromosome 5, 9, and 15 in the MM FISH panel [
20,
22]. Here, we propose a FISH panel for adult B-lineage ALL; a FISH panel for children was not considered in this study.
In contrast to ALL and MM, an appropriate strategy should be selected in AML and MDS, depending on the results of G-banded karyotype. We recommend routine FISH analysis in cases with a complex karyotype, because FISH can identify details of aberrations that cannot be resolved by G-banded karyotype alone. In addition, in cases with few or no mitotic cells, routine FISH analysis would be of benefit for obtaining clinically relevant information regarding aberrations. In AML or MDS with a normal karyotype, certain specific probes could be used to detect cryptic aberrations undetectable by G-banded karyotype. Among FISH probes that we did not evaluate in the present study, the
TP53 probe may be needed in cases with complex karyotypes or insufficient metaphases; this is because the
TP53 deletion is known to be associated with a poor prognosis [
26,
27].
Of particular relevance, FISH analysis may be a superior method for disease monitoring, considering that G-banded karyotype could be hampered by a low in vitro mitotic activity of cancer cells after treatment. Therefore, if specific chromosomal abnormalities are detected by G-banded karyotype at diagnosis, FISH analysis using target probes corresponding to the specific chromosomal abnormalities is needed to investigate specific abnormal FISH signal patterns for use of monitoring markers during follow-up. An advantage of FISH over G-banded karyotype is that it requires considerably less time and effort. Therefore, apart from our proposal, FISH using specific target probes could be utilized in the initial assessment as an adjunct to G-banded karyotype, when critical genetic aberrations (such as PML/RARA rearrangement) should be rapidly identified to determine the best therapeutic approaches.
In addition to G-banded karyotype and FISH, PCR techniques are currently being employed. Of particular interest, multiplex reverse-transcription PCR can be used to simultaneously identify numerous different translocations or chromosomal rearrangements. To detect fusion transcripts or translocations, either PCR or FISH could be used. However, PCR techniques are unable to identify deletions or amplifications that can be readily identified by FISH, and therefore it is unlikely that these techniques will serve as a substitute for FISH. Further study is needed to develop efficient and cost-effective strategies that combine the use of G-banded karyotype, FISH, and PCR techniques.
In conclusion, this study suggests that in the setting of an adequate karyotype of AML and MDS, routine FISH testing contributes little, if any, further genetic information. In contrast, FISH panel testing for ALL and MM appears to be an efficient screening method, and routine FISH analysis should remain the method of choice. Finally, a consensus needs to be reached between laboratories as to the practical strategies for cost-effective utilization of FISH combined with G-banded karyotype.
Go to :






 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download