Abstract
Background
For patients with multiple myeloma (MM), different strategies are used to detect chromosomal abnormalities (CA). There have been a few studies that have directly compared FISH with conventional cytogenetics (CC) for the detection of CA. In this study, we employed a combined approach of metaphase cytogenetics and interphase FISH to investigate the genetic basis for the great heterogeneity observed in the clinical behavior of 28 MM patients.
Methods
Cytogenetic analysis was performed via traditional metaphase karyotype analysis. The FISH studies were done using DNA probes to detect translocations involving the immunoglobulin heavy chain gene (IGH) at 14q32 and deletions of 17p13.1 and 13q14.
Results
CA were detected by CC in 16 patients (57.1%) and by FISH in 14 patients (50.0%) of the 28 patients we studied. 14q32 abnormalities and deletion abnormalities of 13q14 and 17p13.1 were detected by CC in five patients (17.9%), three patients (10.7%) and no patients (0%), respectively and these were detected by FISH in 12 (42.8%), four (14.3%) and five (17.8%), respectively, of the 28 patients we studied. The median follow-up timefor the patients was 23.85 months (range: 0.3∼58.13 months). On the univariate and multivariate analyses, none of the abnormalities detected by cytogenetics and interphase FISH affected survival.
References
2. Seong C, Delasalle K, Hayes K, et al. Prognostic value of cytogenetics in multiple myeloma. Br J Haematol. 1998; 101:189–94.

3. Bergsagel PL, Kuehl WM. Chromosome translocations in multiple myeloma. Oncogene. 2001; 20:5611–22.

4. Kaufmann H, Krömer E, Nösslinger T, et al. Both chromosome 13 abnormalities by metaphase cytogenetics and deletion of 13q by interphase FISH only are prognostically relevant in multiple myeloma. Eur J Haematol. 2003; 71:179–83.

5. Fonseca R, Blood E, Rue M, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003; 101:4569–75.

6. Drach J, Ackermann J, Fritz E, et al. Presence of a p53 gene deletion in patients with multiple myeloma predicts for short survival after conventional-dose chemotherapy. Blood. 1998; 92:802–9.

7. Sawyer JR, Waldron JA, Jagannath S, Barlogie B. Cytogenetic findings in 200 patients with multiple myeloma. Cancer Genet Cytogenet. 1995; 82:41–9.

8. Chang H, Li D, Zhuang L, et al. Detection of chromosome 13q deletions and IgH translocations in patients with multiple myeloma by FISH: comparison with karyotype analysis. Leuk Lymphoma. 2004; 45:965–9.

9. Dewald GW, Therneau T, Larson D, et al. Relationship of patient survival and chromosome anomalies detected in metaphase and/or interphase cells at diagnosis of myeloma. Blood. 2005; 106:3553–8.

10. Gertz MA, Lacy MQ, Dispenzieri A, et al. Clinical implications of t(11;14)(q13;q32), t(4;14)(p16.3;q32), and −17p13 in myeloma patients treated with high-dose therapy. Blood. 2005; 106:2837–40.

11. Shaughnessy J Jr, Tian E, Sawyer J, et al. Prognostic impact of cytogenetic and interphase fluorescence in situ hybridization-defined chromosome 13 deletion in multiple myeloma: early results of total therapy II. Br J Haematol. 2003; 120:44–52.

12. American College of Medical Genetics. Standards and guidelines for clinical genetics laboratories. 2006 Edition.
13. Shaffer L, Tommerup N. An international system for human cytogenetic nomenclature. Basel, Switzerland: S. Karger, 2005. and haemopoietic stem cell transplantation. Myeloma Subcommittee of the EBMT. European Group for Blood and Marrow Transplant. Br J Haematol. 1998; 102:1115–23.
14. BladéJ, Samson D, Reece D, et al. Criteria for evaluating disease response and progression in patients with multiple myeloma treated by high-dose therapy.
15. Lai JL, Zandecki M, Mary JY, et al. Improved cytogenetics in multiple myeloma: a study of 151 patients including 117 patients at diagnosis. Blood. 1995; 85:2490–7.

16. Tabernero D, San Miguel JF, Garcia-Sanz M, et al. Incidence of chromosome numerical changes in multiple myeloma: fluorescence in situ hybridization analysis using 15 chromosome-specific probes. Am J Pathol. 1996; 149:153–61.
17. Drach J, Schuster J, Nowotny H, et al. Multiple myeloma: high incidence of chromosomal aneuploidy as detected by interphase fluorescence in situ hybridization. Cancer Res. 1995; 55:3854–9.
18. Nishida K, Tamura A, Nakazawa N, et al. The Ig heavy chain gene is frequently involved in chromosomal translocations in multiple myeloma and plasma cell leukemia as detected by in situ hybridization. Blood. 1997; 90:526–34.

19. Fonseca R, Barlogie B, Bataille R, et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 2004; 64:1546–58.
20. Zojer N, Königsberg R, Ackermann J, et al. Deletion of 13q14 remains an independent adverse prognostic variable in multiple myeloma despite its frequent detection by interphase fluorescence in situ hybridization. Blood. 2000; 95:1925–30.

21. Avet-Loiseau H, Li JY, Godon C, et al. P53 deletion is not a frequent event in multiple myeloma. Br J Haematol. 1999; 106:717–9.

22. Schultheis B, Krämer A, Willer A, Hegenbart U, Goldschmidt H, Hehlmann R. Analysis of p73 and p53 gene deletions in multiple myeloma. Leukemia. 1999; 13:2099–103.

23. Mazars GR, Portier M, Zhang XG, et al. Mutations of the p53 gene in human myeloma cell lines. Oncogene. 1992; 7:1015–8.
Fig. 1.
Interphase fluorescence in situ hybridization (FISH) findings. (A) A plasma cell shows rearrangement of IGH. (B) A plasma cell shows deletion 13q14. The locus-specific 13q14 probes are labeled by orange color. (C) A plasma cell shows deletion 17p13.1. The locus-specific p53 probes are labeled by orange color.
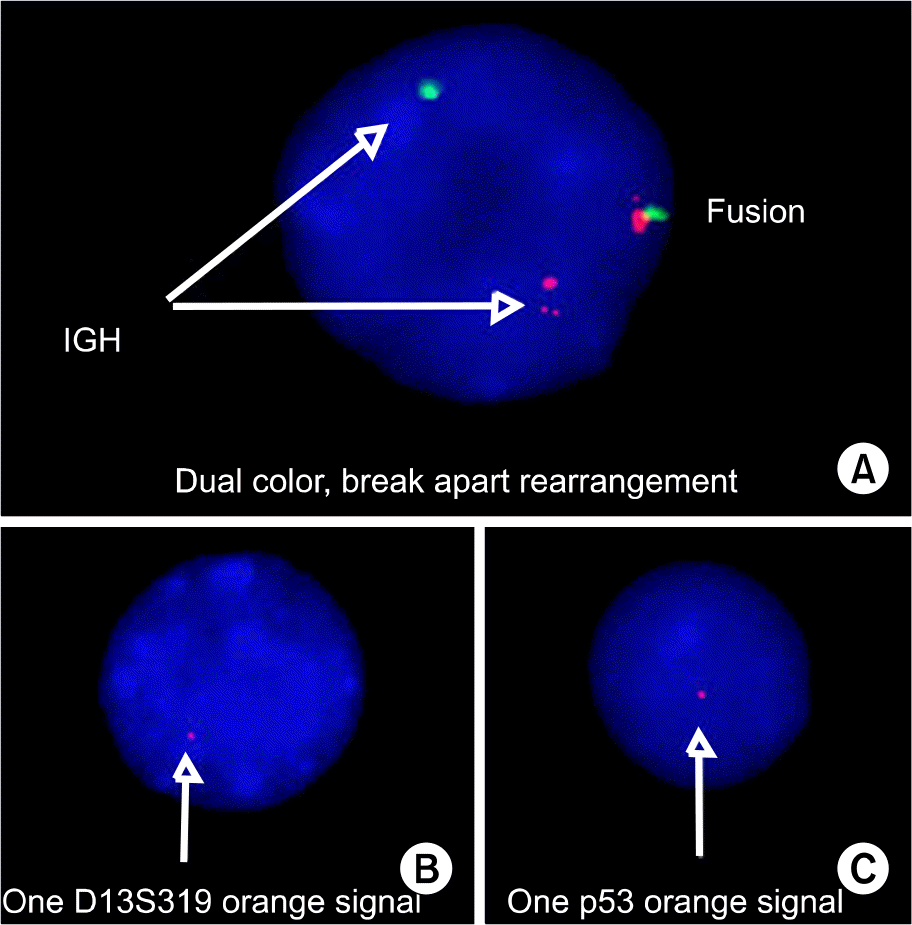
Fig. 2.
The overall survival curves according to the initial treatment response. For patients with a CR or a PR, the median survival has not yet been reached. The median survival was 7.03 months for patients with MR. There were significant differences in survival according to the initial treatment response (P=0.036). (CR; complete response, PR; partial response, MR; minimal response).
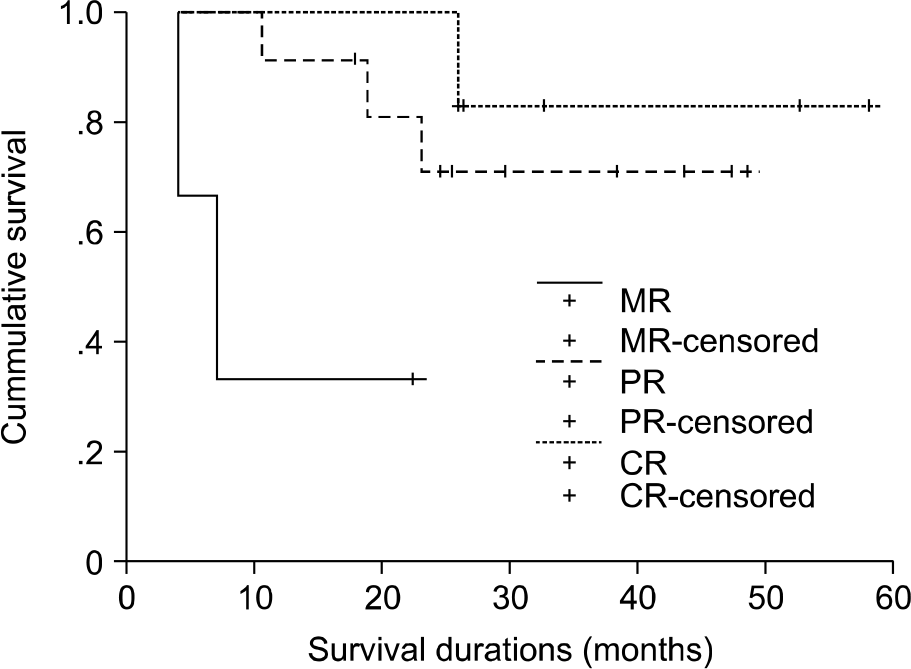
Table 1.
Characteristics of the 28 study patients
Table 2.
CC and FISH data from 28 MM patients
Table 3.
P value for the association between the bioclinical features and the abnormalities n=28




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download