Abstract
Background:
The FUT2 and FUT3 genes determine the Lewis phenotype of red blood cells (RBCs). Recently, the Lewis genes, the secretor genes, and several mutations that cause Lewis negative and nonsecretor phenotypes have been identified. The purpose of this study was to analyze the gene frequency of FUT2 and FUT3 in a Korean population by comparing the use of the direct sequencing method to the use of the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method for mutation detection in the FUT2 and FUT3 genes.
Methods:
RBCs and peripheral blood leukocytes were obtained from 225 apparently healthy volunteers to determine the phenotype and genotype of the FUT2 and FUT3 genes. Lewis phenotypes were determined on K3EDTA-stablized fresh blood samples using the column agglutination method. Lewis blood group genotyping was performed by use of the direct sequencing method. For the detection of T59G, C357T, and A385T mutations, the PCR-RFLP method was performed.
Results:
The frequencies of the Lewis blood group phenotype were 12.4% for Le(a+b−), 70.7% for Le(a−b+), 11.1% for Le(a−b−) and 5.8% for Le(a+b+), respectively. Direct Sequencing of the FUT2 gene identified 92.2% C357T, 56.9% A385T, 0.4% G244A mutations and 1.8% del396. Direct Sequencing of the FUT3 gene identified 46.9% T59G, 30.4% G508A, 1.1% T202C, 1.1% C314T, 0.7% A1029G, 3.0% T1067A and 13.3% G1242A mutations. The PCR-RFLP method results were discordant in nine cases (1 case for C357T, 4 cases for A385T and 2 cases for T59G) as compared to the direct sequencing method results.
Go to : 
REFERENCES
1). McCurley RS., Recinos A 3rd., Olsen AS, et al. Physical maps of human alpha(1,3)fucosyltransferase genes FUT3-FUT6 on chromosomes 19p13.3 and 11q21. Genomics. 1995. 26:142–6.
2). Rouquier S., Lowe JB., Kelly RJ., Fertitta AL., Lennon GG., Giorgi D. Molecular cloning of a human genomic region containing the H blood group alpha(1,2)fucosyltransferase gene and two H locus-related DNA restriction fragments. Isolation of a candidate for the human Secretor blood group locus. J Biol Chem. 1995. 270:4632–9.
3). Hammer L., Månsson S., Rohr T, et al. lewis phenotype of erythrocytes and Leb-active actine glycolipid in serum of pregnant women. Vox Sang. 1981. 40:27–33.
4). Stigendal L., Olsson R., Rydberg L., Samuelsson BE. Blood group Lewis phenotype on erythrocytes and in saliva in alcoholic pancreatitis and chronic liver disease. J Clin Pathol. 1984. 37:778–82.

5). Liu TC., Chang JG., Lin SF, et al. Lewis (FUT3) genotypes in Taiwanese, Thai, and Filipino populations. Ann Hematol. 2000. 79:599–603.

6). Larson G., Svensson L., Hynsjӧ L., Elmgren A., Rydberg L. Typing for the human lewis blood group system by quantitative fluorescence-activated flow cytometry: large differences in antigen presentation on erythrocytes between A(1), A(2), B, O phenotypes. Vox Sang. 1999. 77:227–36.

7). Chang JG., Yang TY., Liu TC, et al. Molecular analysis of secretor type alpha(1,2)-fucosyltransferase gene mutations in the Chinese and Thai populations. Transfusion. 1999. 39:1013–7.

8). Costache M., Cailleau A., Fernandez-Mateos P., Oriol R., Mollicone R. Advances in molecular genetics of alpha-2- and alpha-3/4-fucosyltransferases. Transfus Clin Biol. 1997. 4:367–82.
9). Serpa J., Almeida R., Oliveira C, et al. Lewis enzyme (alpha1-3/4 fucosyltransferase) polymorphisms do not explain the Lewis phenotype in the gastric mucosa of a Portuguese population. J Hum Genet. 2003. 48:183–9.
10). Ogilvie EM., Fife MS., Thompson SD, et al. The -174G allele of the interleukin-6 gene confers susceptibility to systemic arthritis in children: a multi-center study using simplex and multiplex juvenile idiopathic arthritis families. Arthritis Rheum. 2003. 48:3202–6.

11). Buono P., Pasanisi F., Nardelli C, et al. Six novel mutations in the proopiomelanocortin and melano-cortin receptor 4 genes in severely obese adults living in southern Italy. Clin Chem. 2005. 51:1358–64.

12). Andersson B., Ying JH., Lewis DE., Gibbs RA. Rapid characterization of HIV-1 sequence diversity using denaturing gradient gel electrophoresis and direct automated DNA sequencing of PCR products. PCR Methods Appl. 1993. 2:293–300.

13). Suh IB., Kim YK. Relationship of Helicobacter pylori IgG seroprevalence with Lewis (a,b) blood group phenotype/genotype. Korean J Blood Transfus. 2001. 12:35–45.
14). Napierala D., Garcia-Rojas X., Sam K, et al. Mutations and promoter SNPs in RUNX2, a transcriptional regulator of bone formation. Mol Genet Metab. 2005. 86:257–68.

15). Cowles JW., Cox A., McMican A., Blumberg N. Comparison of monoclonal antisera with conventional antisera for Lewis blood group antigen determination. Vox Sang. 1987. 52:83–4.

16). Young WW Jr., Johnson HS., Tamura Y, et al. Characterization of monoclonal antibodies specific for the Lewis a human blood group determinant. J Biol Chem. 1983. 25: 258:4890–4.

17). Rouger P., Noizat-Pienne F., Le Pennec PY. Advances in the use of monoclonal antibodies for blood group testing. Transfus Clin Biol. 1997. 4:345–9.

18). Good AH., Yau O., Lamontagne LR., Oriol R. Serological and chemical specificities of twelve monoclonal anti-Lea and anti-Leb antibodies. Vox Sang. 1992. 62:180–9.
19). Ameno S., Ameno K., Kinoshita H., Tanaka N., Ijiri I. Lewis genotyping by the PCR-RFLP method in a Japanese population and its evaluation in forensic analysis. Int J Legal Med. 1997. 110:232–4.

20). Nishihara S., Narimatsu H., Iwaski H, et al. Molecular genetic analysis of the human Lewis histo-blood group system. J Biol Chem. 1994. 269:29271–8.

21). Yazawa S., Oh-kawara H., Nakajima T., Hosomi O., Akamatsu S., Kishi K. Histo-blood group Lewis genotyping from human hairs and blood. Jpn J Hum Genet. 1996. 41:177–88.

22). Kukowska-Latallo JF., Larsen RD., Nair RP., Lowe JB. A cloned human cDNA determines expression of a mouse stage-specific embryonic antigen and the Lewis blood group alpha(1,3/1,4)fucosyltransferase. Genes Dev. 1990. 4:1288–303.

23). Kelly RJ., Rouquier S., Giorgi D., Lennon GG., Lowe JB. Sequence and expression of a candidate for the human secretor blood group alpha(1,2)fucosyltrans-ferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secrector phenotype. J Biol Chem. 1995. 270:4640–9.
24). Kudo T., Iwasaki H., Nishihara S, et al. Molecular genetic analysis of the human Lewis histo-bood group system. II. Secrector gene inactivation by a novel single missense mutation A385T in Japanese nonsecretor individuals. J Biol Chem. 1996. 271:9830–7.
Go to : 
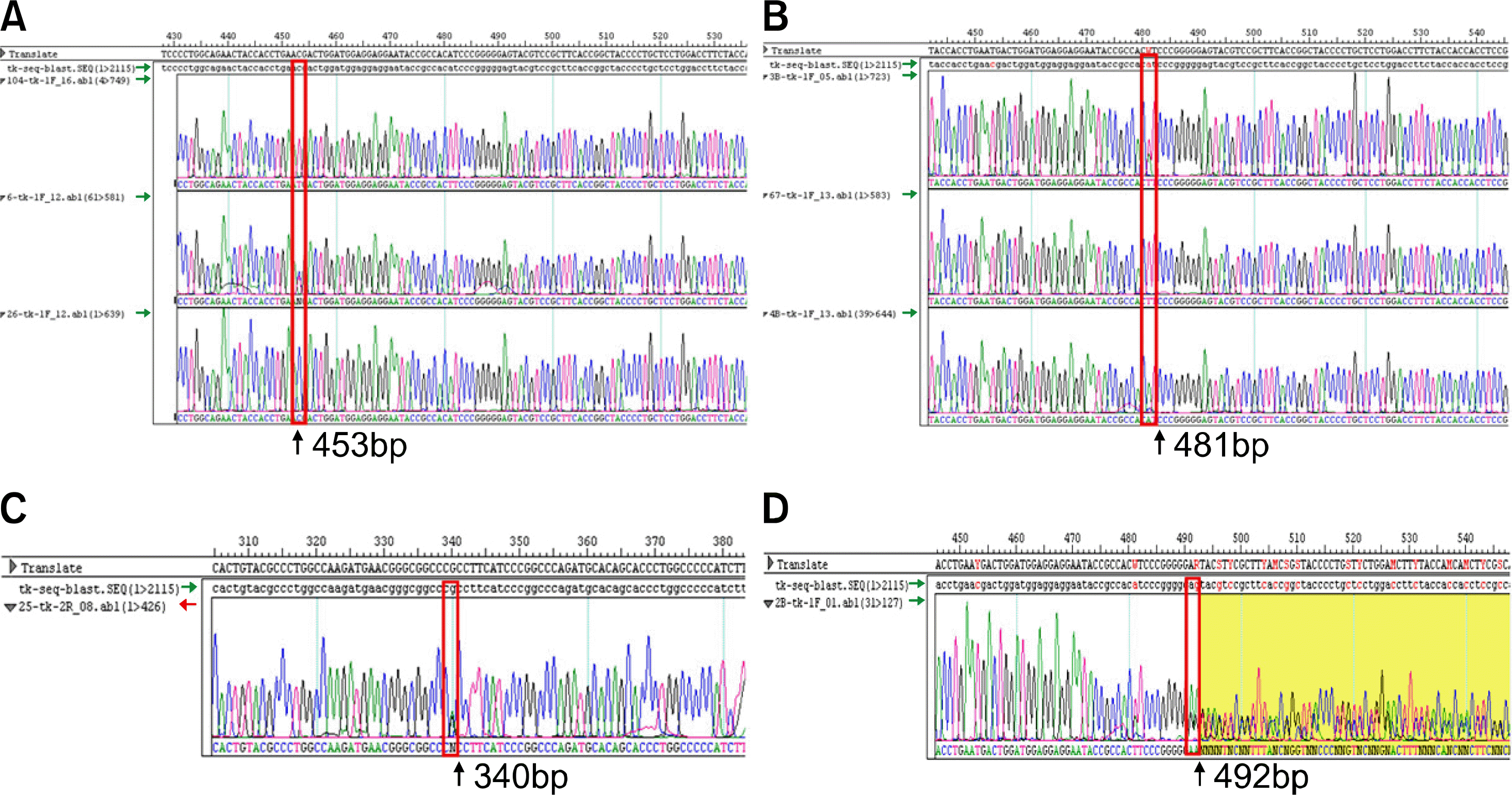 | Fig. 1Chromatograms of direct sequencing for FUT2 gene mutations. (A) C357T heterozygote mutant. (B) G385A heterozygote mutant. (C) G244A wild type. (D) del396 mutant. |
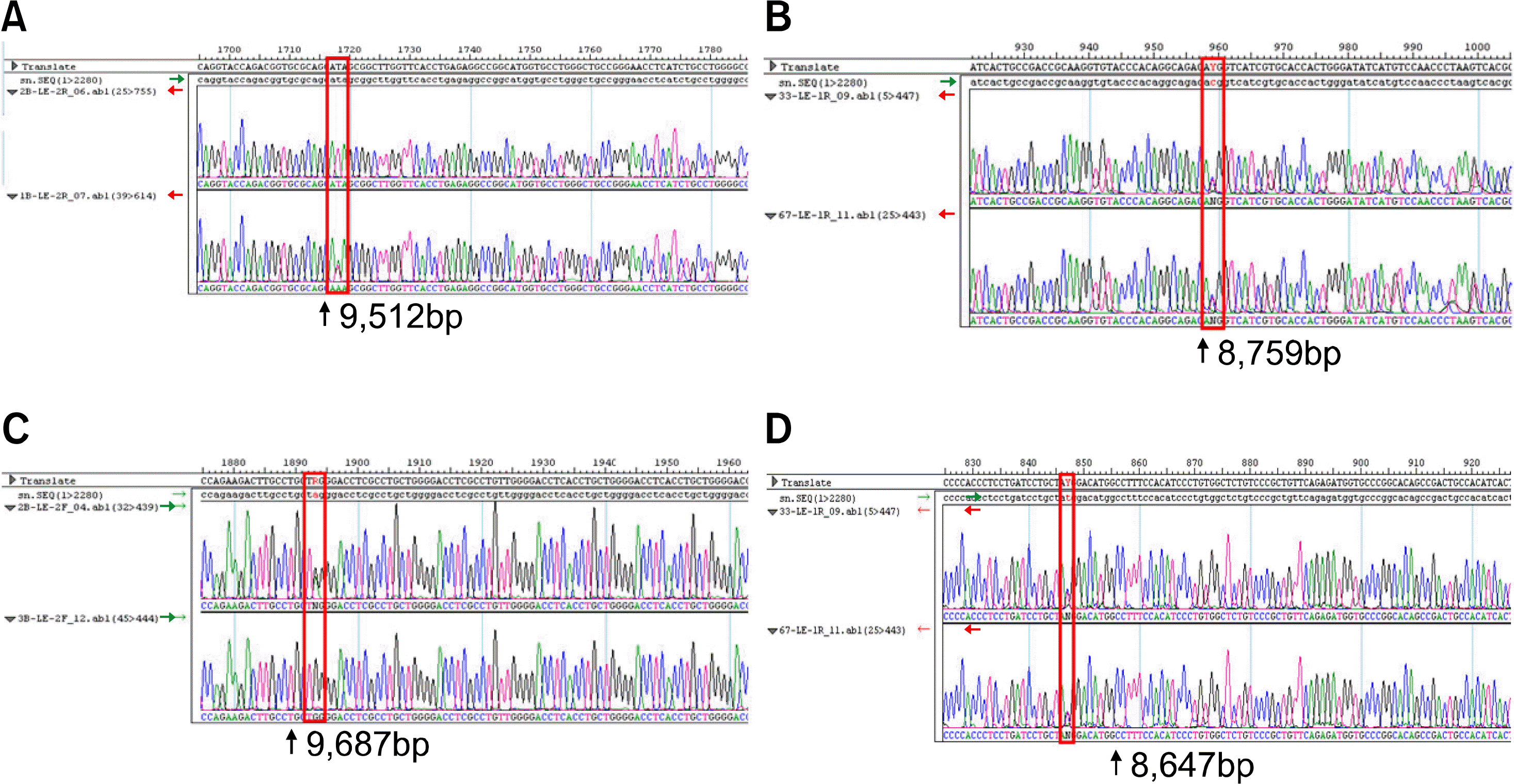 | Fig. 2Chromatograms of direct sequencing for FUT3 gene mutations. (A) T1067A heterozygote mutant. (B) T202C heterozygote mutant. (C) G1242A heterozygote mutant. (D) C314T heterozygote mutant. |
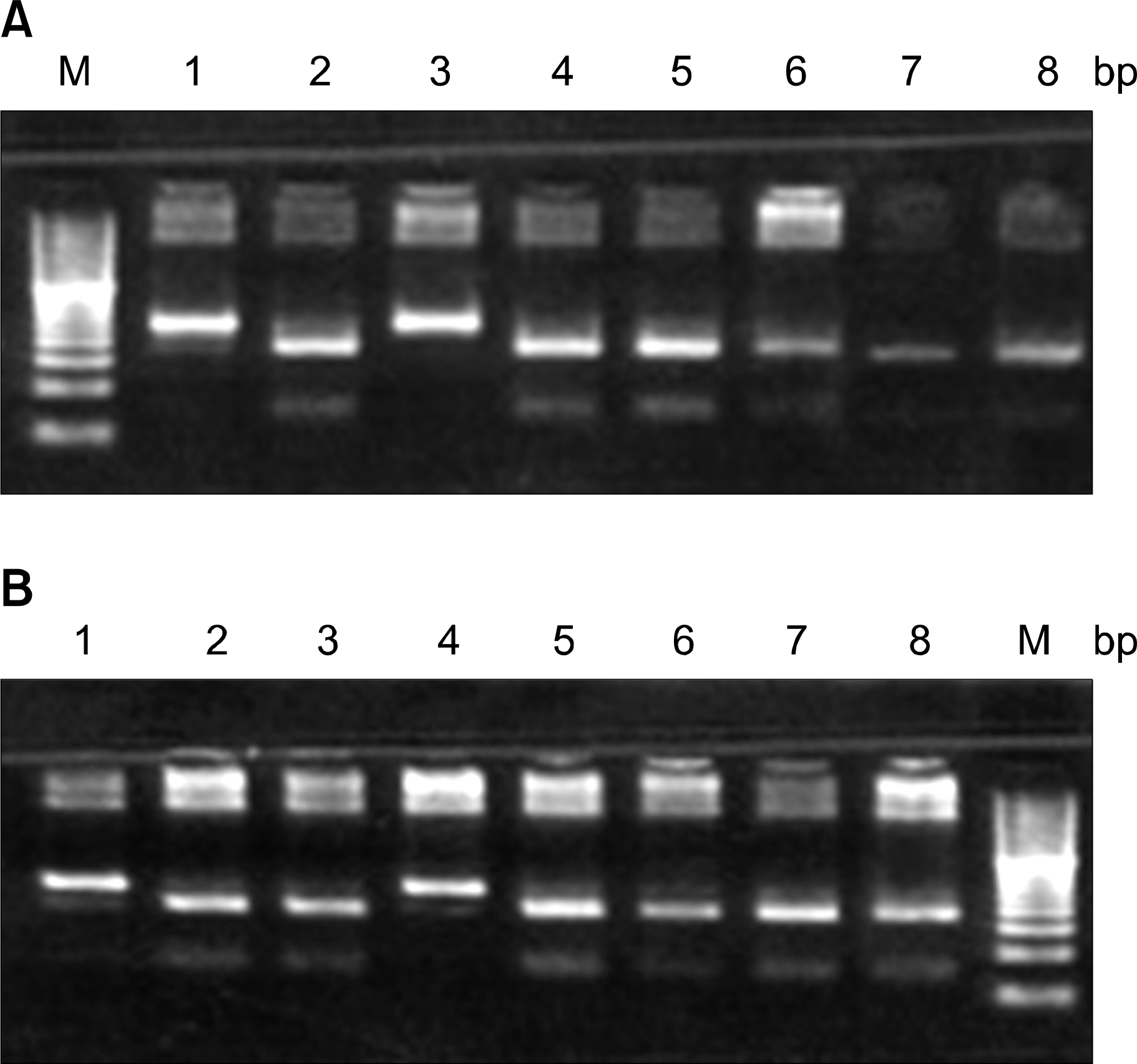 | Fig. 3(A) Electrophoresis pattern for the detection of FUT2 gene mutation (C357T) using AseI restriction enzyme. Lane M-20bp DNA ladder; lane 1,3-wild type; lane 2,4,5,6,7,8het-erozygote mutants. (B) Electrophoresis pattern for the detection of FUT2 gene mutation (A385T) using AluI restriction enzyme. Lane M-20bp DNA ladder; lane 1,4-wild type; lane 2,3,5,6,7,8-heterozygote mutants. |
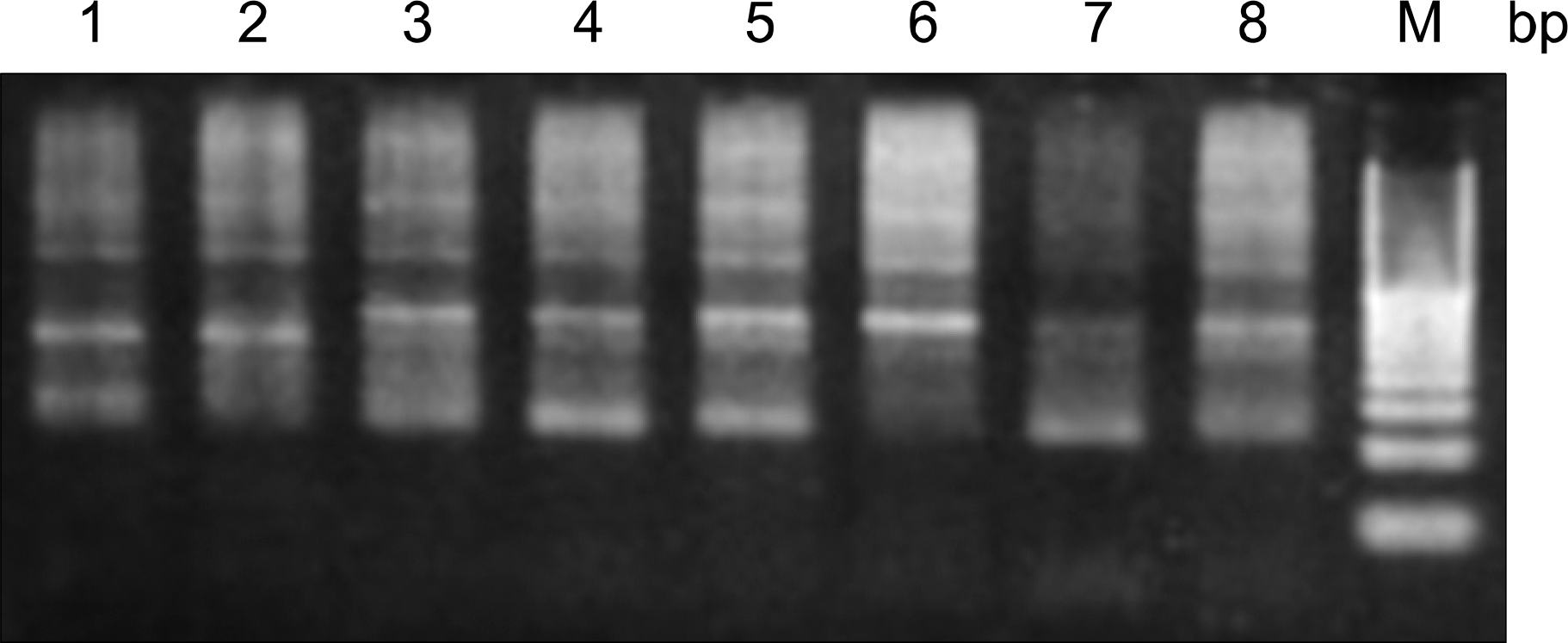 | Fig. 4Electrophoresis pattern for the detection of FUT3 gene mutation (T59G) using MspI restriction enzyme. Lane M-20bp DNA ladder; lane 6-wild type; lane 1,3,4,5,8-heterozygote mutants; lane 7-homozygotic mutants. |
Table 1.
Primers and PCR conditions in FUT2 and FUT3 gene
Table 2.
Primers for nested PCR and direct sequencing in FUT2 and FUT3 gene
Table 3.
PCR primers of the nested PCR reaction for the RFLP analysis of FUT2 and FUT3 gene
Table 4.
Restriction enzymes for the detection of FUT2 and FUT3 gene mutations
Table 5.
The distribution of ABO blood group and Lewis phenotype
Table 6.
The frequencies of FUT2 and FUT3 base substitutions in Korean




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download