Abstract
Background:
Immunoglobulin heavy chain (IgH) gene rearrangement has been known to be a useful marker for determining the clonality as well as detecting minimal residual disease in B cell malignancies. This study was performed to establish single polymerase chain reaction (PCR) methods for the detection of IgH gene rearrangements in formalin-fixed, paraffin-embedded tissue of patients with B cell lymphoma and determine the type of JH segments used.
Methods:
We obtained formalin-fixed, paraffin-embedded tissue sections of 44 patients diagnosed with B cell lymphoma at Ajou University Hospital from January 2005 to January 2007 and reviewed medical records retrospectively. After the extraction of DNA, PCR was performed using VH3 and JHPST primers to detect the third complementarity determining region (CDR3) gene of IgH. Sequence analysis of the PCR products was also done in 23 patients.
Results:
The CDR3 gene rearrangements were detected in 26 (59%) out of 44 patients with B cell lymphoma. Sequence analysis of the amplified CDR3 gene was successful in 16 (70%) of 23 patients. JH3, JH4, JH5, and JH6 segments were used for CDR3 gene rearrangements in 3 (25%), 4 (33%), 1 (8%), and 4 (33%) patients with diffuse large B cell lymphoma, respectively.
Conclusion:
Although there are some limitations due to a low sensitivity less than 60%, single PCR using consensus primers could be an effective tool for the detection of CDR3 gene rearrangements in routine laboratory settings. Furthermore, sequence analysis of the CDR3 PCR products will provide basic information necessary for further studies.
Go to : 
REFERENCES
1). van Dongen JJ., Wolvers-Tettero IL. Analysis of immunoglobulin and T cell receptor genes. Part II: possibilities and limitations in the diagnosis and management of lymphoproliferative diseases and related disorders. Clin Chim Acta. 1991. 198:93–174.
2). Early P., Huang H., Davis M., Calame K., Hood L. An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH, D and JH. Cell. 1980. 19:981–92.

4). Yamada M., Hudson S., Tournay O, et al. Detection of minimal disease in hematopoietic malignancies of the B-cell lineage by using third-complementarity-determining region (CDR-III)-specific probes. Proc Natl Acad Sci U S A. 1989. 86:5123–7.

5). van Dongen JJ., Wolvers-Tettero IL. Analysis of immunoglobulin and T cell receptor genes. Part I: basic and technical aspects. Clin Chim Acta. 1991. 198:1–91.
6). van Dongen JJ., Langerak AW., Bruggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003. 17:2257–317.

7). Lehmann U., Kreipe H. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods. 2001. 25:409–18.

8). Apostolidis J., Gupta RK., Grenzelias D, et al. High-dose therapy with autologous bone marrow support as consolidation of remission in follicular lymphoma: long-term clinical and molecular follow-up. J Clin Oncol. 2000. 18:527–36.

9). Gribben JG., Neuberg D., Freedman AS, et al. Detection by polymerase chain reaction of residual cells with the bcl2 translocation is associated with increased risk of relapse after autologous bone marrow transplantation for B-cell lymphoma. Blood. 1993. 81:3449–57.

10). Moos M., Schulz R., Martin S., Benner A., Haas R. The remission status before and the PCR status after high-dose therapy with peripheral blood stem cell support are prognostic factors for relapse-free survival in patients with follicular non-Hodgkin's lymphoma. Leukemia. 1998. 12:1971–6.

11). Zwicky CS., Maddocks AB., Andersen N., Gribben JG. Eradication of polymerase chain reaction detectable immunoglobulin gene rearrangement in non-Hodg-kin's lymphoma is associated with decreased relapse after autologous bone marrow transplantation. Blood. 1996. 88:3314–22.

12). Park CJ., Kim MC., Moon AR, et al. Detection of minimal residual disease by IgH gene rearrangement-PCR in childhood acute lymphoblastic leukemia. Korean J Clin Pathol. 1999. 19:163–71.
13). Cho EY., Ko YH., Kim DS., Han JJ., Ree HJ. Diagnostic utility of polymerase chain reaction-based clonality analysis for immunoglobulin heavy chain gene and T-cell receptor gamma chain gene rearrangement in lymphoid neoplasms. Korean J Pathol. 2001. 35:461–9.
14). Bagg A., Braziel RM., Arber DA., Bijwaard KE., Chu AY. Immunoglobulin heavy chain gene analysis in lymphomas: a multi-center study demonstrating the heterogeneity of performance of polymerase chain reaction assays. J Mol Diagn. 2002. 4:81–9.
15). Goelz SE., Hamilton SR., Vogelstein B. Purification of DNA from formaldehyde fixed and paraffin embedded human tissue. Biochem Biophys Res Commun. 1985. 130:118–26.

16). Tierens A., Lozano MD., Wickert R., Chan WC., Greiner TC. High-resolution analysis of immunoglobulin heavy-chain gene rearrangements using denaturing gradient gel electrophoresis. Diagn Mol Pathol. 1996. 5:159–65.

17). Wu G., Greiner TC., Chang WC. Obtaining clone-specific primer and probe for the immunoglobulin heavy-chain gene from paraffin-embedded tissue of B-cell lymphoma: technical considerations. Diagn Mol Pathol. 1997. 6:147–53.
18). Elenitoba-Johnson KS., Bohling SD., Mitchell RS., Brown MS., Robetorye RS. PCR analysis of the immunoglobulin heavy chain gene in polyclonal processes can yield pseudoclonal bands as an artifact of low B cell number. J Mol Diagn. 2000. 2:92–6.

19). Buchonnet G., Jardin F., Jean N, et al. Distribution of BCL2 breakpoints in follicular lymphoma and correlation with clinical features: specific subtypes or same disease? Leukemia. 2002. 16:1852–6.

20). Brezinschek HP., Brezinschek RI., Lipsky PE. Analysis of the heavy chain repertoire of human peripheral B cells using single-cell polymerase chain reaction. J Immunol. 1995. 155:190–202.
21). Welzel N., Le T., Marculescu R, et al. Templated nucleotide addition and immunoglobulin JH-gene utilization in t(11;14) junctions: implications for the mechanism of translocation and the origin of mantle cell lymphoma. Cancer Res. 2001. 61:1629–36.
22). Jager U., Bocskor S., Le T, et al. Follicular lymphomas' BCL-2/IgH junctions contain templated nucleotide insertions: novel insights into the mechanism of t(14;18) translocation. Blood. 2000. 95:3520–9.
Go to : 
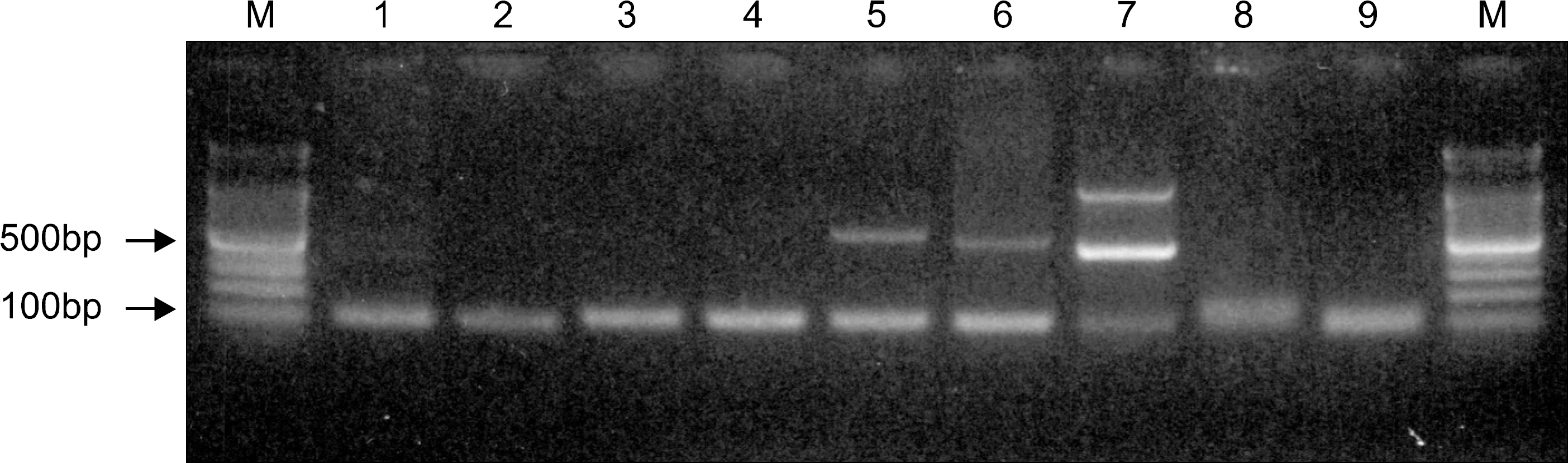 | Fig. 12% agarose gel electrophoresis of the PCR product amplified for CDR3 gene. M, size marker (100bp ladder); 1, positive control; 2∼4, positive samples; 5∼7, positive samples with one or more nonspecific bands; 8, negative control; 9, negative sample. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download