Abstract
Background:
The global effect of HIV infection on the host cell gene expression profiles in healthy HIV-infected patients, as long-term non-progressors, remains largely unknown. To identify the cellular genes related with HIV infection and delayed disease progression in vivo, the host gene expression profiles between healthy HIV-infected Koreans and AIDS patients were investigated.
Methods:
Differential expression gene analysis was performed via oligonucleotide microarray with using Magic-oligo 10K chip. Ten HIV-uninfected persons and 10 HIV-infected patients (healthy HIV-infected patients vs. AIDS patients. respectively) were studied.
Results:
Only 10.8% (1,097 genes) of the total genes, that is, 331 up-regulated genes and 766 down-regulated genes were differentially expressed with more than a two-fold change in the HIV-infected persons as compared to those of the HIV-uninfected persons. Especially, 97 genes (8.8%) among 1,097 genes were commonly up- or down-regulated in both the healthy HIV-infected patients and the AIDS patients. 187 genes were differently expressed on the gene expression analysis between the healthy HIV-infected patients and the AIDS patients. Twenty-eight genes out of them showed very significant differences with a P value <0.01. Especially, tripartite motif (TRIM) 14 protein and interferon gamma receptor 2 were dramatically up-regulated in healthy HIV-infected patients, while death-associated protein, DNA directed RNA polymerase II polypeptide A and STAT were overexpressed in AIDS patients.
Go to : 
REFERENCES
1). Geiss GK., Bumgarner RE., An MC, et al. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology. 2000. 266:8–16.

2). Swingler S., Mann A., Jacque J, et al. HIV-1 Nef mediates lymphocyte chemotaxis and activation by infected macrophages. Nat Med. 1999. 5:997–1003.

3). Izmailova E., Bertley FM., Huang Q, et al. HIV-1 Tat reprograms immature dendritic cells to express chemoattractants for activated T cells and macrophages. Nat Med. 2003. 9:191–7.

4). de la Fuente C., Santiago F., Deng L, et al. Gene expression profile of HIV-1 Tat expressing cells: a close interplay between proliferative and differentiation signals. BMC Biochem. 2002. 3:14.
5). Chun TW., Fauci AS. Latent reservoirs of HIV: obstacles to the eradication of virus. Proc Natl Acad Sci U S A. 1999. 96:10958–61.

6). Chun TW., Justement JS., Lempicki RA, et al. Gene expression and viral prodution in latently infected, resting CD4+ T cells in viremic versus aviremic HIV-infected individuals. Proc Natl Acad Sci U S A. 2003. 100:1908–13.

7). Almeida CA., Price P., French MA. Immune activation in patients infected with HIV type 1 and maintaining suppression of viral replication by highly active antiretroviral therapy. AIDS Res Hum Retroviruses. 2002. 18:1351–5.
8). Wilson SA., Sieiro-Vazquez C., Edwards NJ, et al. Cloning and characterization of hIF2, a human homologue of bacterial translation initiation factor 2, and its interaction with HIV-1 matrix. Biochem J. 1999. 342(Pt 1):97–103.

9). Endo-Munoz L., Warby T., Harrich D., McMillan NA. Phosphorylation of HIV Tat by PKR increases interaction with TAR RNA and enhances transcription. Virol J. 2005. 2:17.
10). Chen W., Tang Z., Fortina P, et al. Ethanol potentiates HIV-1 gp120-induced apoptosis in human neurons via both the death receptor and NMDA receptor pathways. Virology. 2005. 334:59–73.

11). Cimarelli A., Luban J. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J Virol. 1999. 73:5388–401.

12). Perales C., Carrasco L., Ventoso I. Cleavage of eIF4G by HIV-1 protease: effects on translation. FEBS Lett. 2003. 533:89–94.

13). Szabo J., Cervenak L., Toth FD, et al. Soluble gC1q-R/p33, a cell protein that binds to the globular “heads” of C1q, effectively inhibits the growth of HIV-1 strains in cell cultures. Clin Immunol. 2001. 99:222–31.
14). van't Wout AB., Lehrman GK., Mikheeva SA, et al. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4(+)-T-cell lines. J Virol. 2003. 77:1392–402.
15). Shaheduzzaman S., Krishnan V., Petrovic A, et al. Effects of HIV-1 Nef on cellular gene expression profiles. J Biomed Sci. 2002. 9:82–96.

16). Jowett JB., Planelles V., Poon B., Shah NP., Chen ML., Chen IS. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol. 1995. 69:6304–13.

17). Rey O., Canon J., Krogstad P. HIV-1 Gag protein associates with F-actin present in microfilaments. Virology. 1996. 220:530–4.

18). Kawai T., Matsumoto M., Takeda K., Sanjo H., Akira S. ZIP kinase, a novel serine/threonine kinase which mediates apoptosis. Mol Cell Biol. 1998. 18:1642–51.

19). Chun RF., Jeang KT. Requirements for RNA polymerase II carboxyl-terminal domain for activated transcription of human retroviruses human T-cell lymphotropic virus I and HIV-1. J Biol Chem. 1996. 271:27888–94.

20). Miller ED., Smith JA., Lichtinger M., Wang L., Su L. Activation of the signal transducer and activator of transcription 1 signaling pathway in thymocytes from HIV-1-infected human thymus. AIDS. 2003. 17:1269–77.

21). Hottiger MO., Nabel GJ. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J Virol. 1998. 72:8252–6.

23). Song B., Javanbakht H., Perron M., Park DH., Strem-lau M., Sodroski J. Retrovirus restriction by TRIM5-alpha variants from Old World and New World primates. J Virol. 2005. 79:3930–7.
24). Sedger LM., Shows DM., Blanton RA, et al. IFN-gamma mediates a novel antiviral activity through dynamic modulation of TRAIL and TRAIL receptor expression. J Immunol. 1999. 163:920–6.
Go to : 
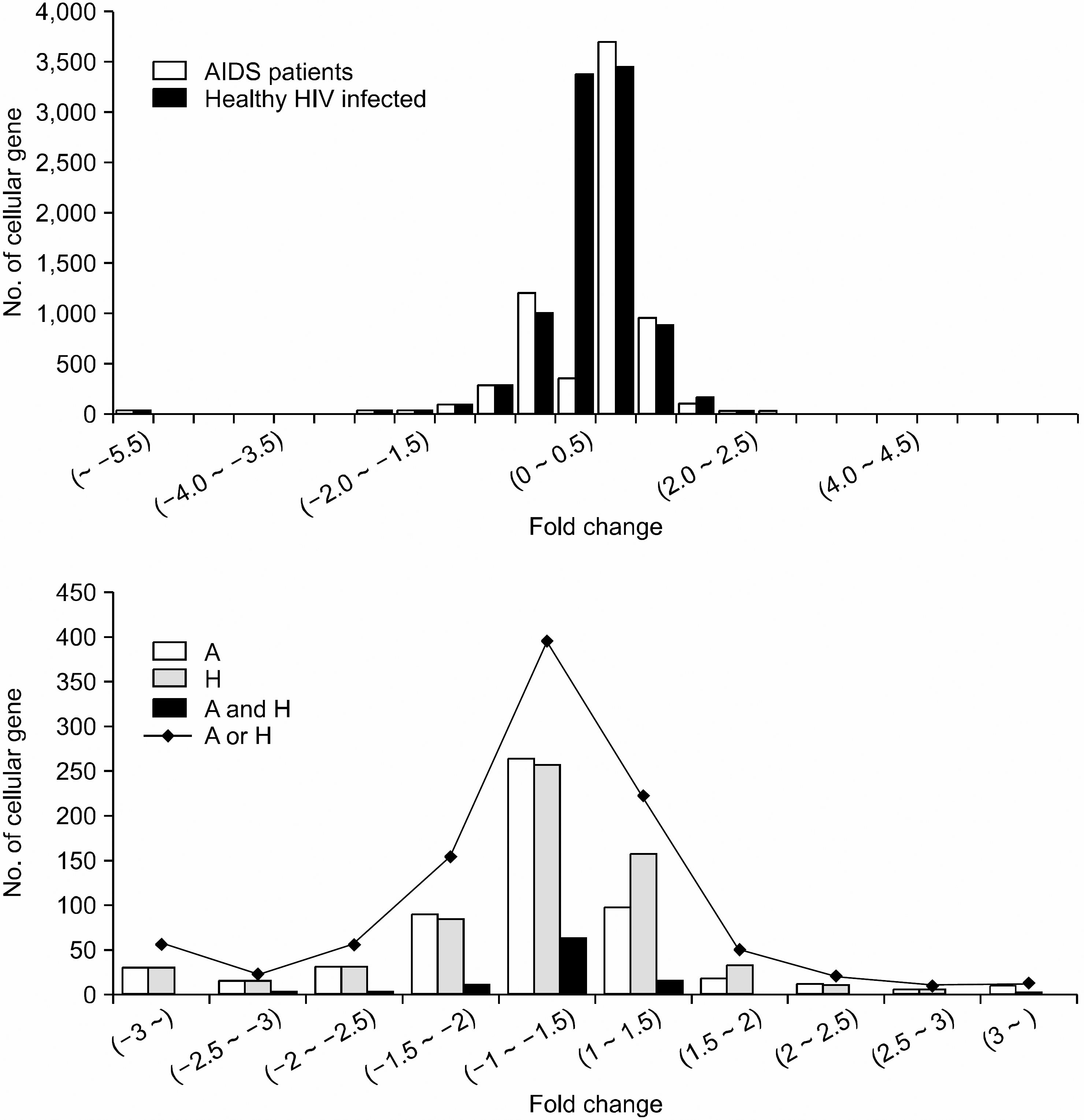 | Fig. 1Distribution of the changes of gene expression profiles in healthy HIV-infected and AIDS patients compared to those of HIV-uninfected persons. The upper and lower show whole gene expression profiles and the distribution of differentially expressed genes (DEGs) with>two-fold change between healthy HIV-infected and AIDS patients, respectively. A and H represent AIDS patients and Healthy HIV-in-fected persons. (A and H) means DEGs that were commonly expressed in two groups and (A or H) means the total number of differentially expressed genes with>two fold change in two groups. Each value of fold change was presented in log 2. |
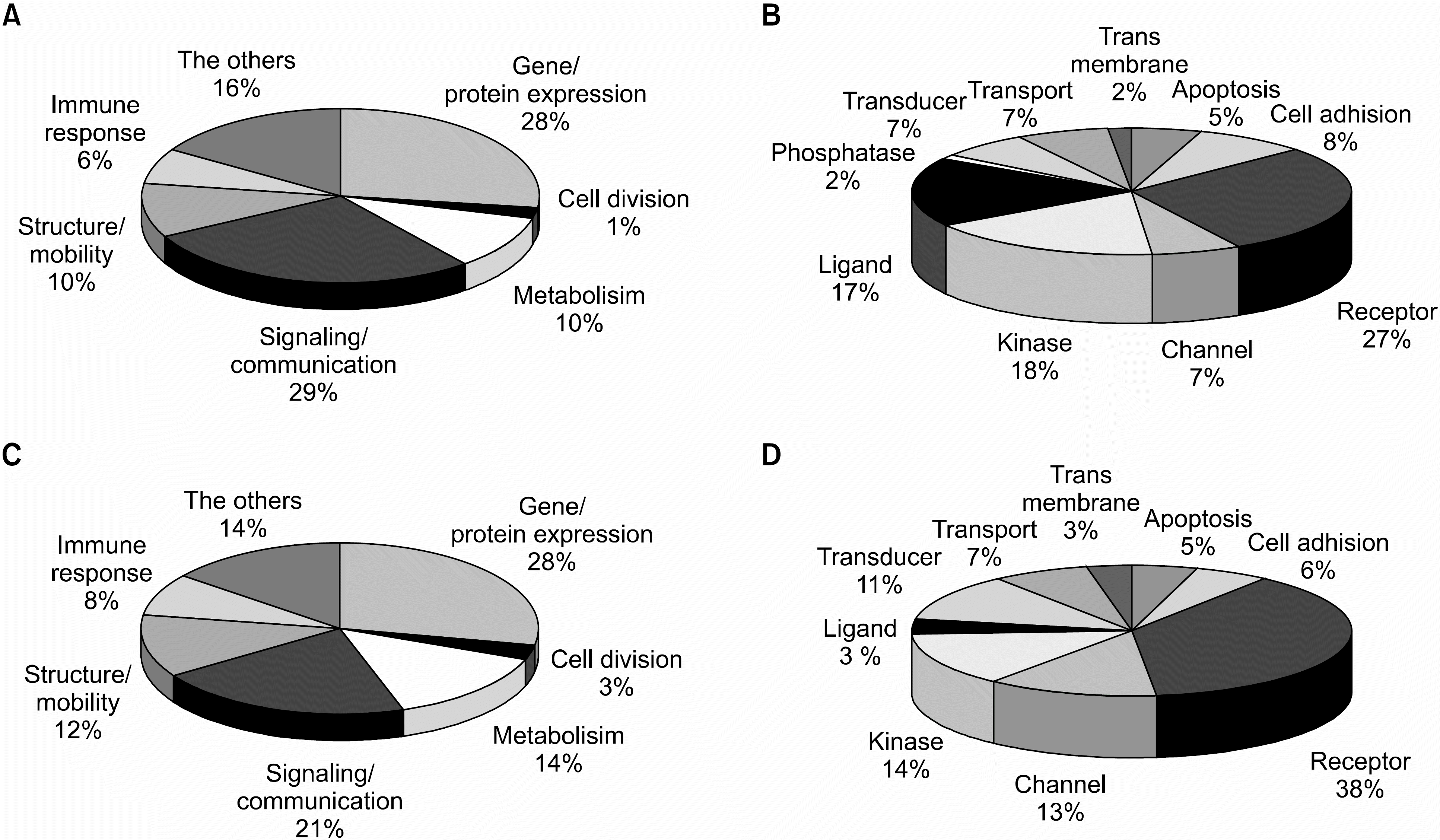 | Fig. 2The functional distribution of differentially expressed genes in HIV-infected patients. DEGs were divided into 7 categories based on their function. Numbers mean the percentages of genes in each category compared to total number of genes regulated. (A) The distribution of 331 up-regulated genes based on their cellular function. Genes that involved in signaling/communication were mostly up-regulated. (B) The distribution of overexpressed signaling/communication related genes in HIV-infected patients according to sub-categories. Signaling/communication genes were further divided into 10 classes. Receptors, ligands, and kinases genes were mostly up-regulated. (C) The distribution of 766 down-regulated genes based on their cellular function. (D) Distribution of down-regulated signaling/communication related genes in HIV-infected patients according to sub-categories. Signaling/communication genes were further divided into 10 classes. Receptors related genes were mostly down-regulated. |
Table 1.
The representative genes that were commonly up-regulated in Healthy HIV-infected and AIDS groups compared to those of HIV-uninfected group
Table 2.
The representative genes that were simultaneously down-regulated in Healthy HIV-infected and AIDS groups compared to those of HIV-uninfected group
Table 3.
Gene lists showing the significant difference with P<0.01 in gene expression profiles between healthy HIV-infected and AIDS patients by the univariate test




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download