Abstract
Background:
In the search for susceptibility genes responsible for leukemia, genetic studies involving HLA association have been in progress extensively since the first report on its effect on the disease. Here we investigated the genetic associations of different leukemias with 4 autosomal mHags, HA-1, -2, -8 and HB-1. In particular, HB-1 is one of the leukemia-associated minor histocompatibility antigens (mHags) that is significantly expressed by Epstein-Barr virus-transformed- and tumor cells of all B lineage acute lymphoblastic leukemia (ALL).
Methods:
A simultaneous genotyping method using PCR sequence-specific primers against HA-1, -2, -8 and HB-1 was developed, and their allelic frequencies in 139 healthy controls and 36 leukemia patients were observed. To compare genotype, phenotype, and gene frequencies of mHags with healthy controls, leukemia patients were classified into sub groups of ALL, acute myeloid leukemia (AML), and chronic myeloid leukemia (CML).
Results:
The genotype frequencies of HA-1, -2 and -8 were not significantly different from healthy controls in every group of leukemia patients. However, the HB-1 H genotype was significantly increased in leukemia patients (P=0.03, OR=1.82, CI=1.08~3.06), particularly in AML (P=0.01, OR=2.4, CI=1.21~ 4.76) as compared with healthy controls.
Conclusion:
Our results suggested that the genotype of HB-1 H may be associated with leukemia, particularly with AML. In further study, it is necessary to confirm the association of HB-1 with other leukemias in a larger group of patients, and to identify the underlying mechanism of HB-1 responsible for the occurrence of leukemia.
Go to : 
REFERENCES
1). Fong CT., Brodeur GM. Down's syndrome and leukemia: epidemiology, genetics, cytogenetics and mechanisms of leukemogenesis. Cancer Genet Cytoge-net. 1987. 28:55–76.

2). Potzsch C., Voigtlander T., Lubbert M. p53 germline mutation in a patient with Li-Fraumeni syndrome and three metachronous malignancies. J Cancer Res Clin Oncol. 2002. 128:456–60.
3). Savitz DA., Andrews KW. Review of epidemiologic evidence on benzene and lymphatic and hematopoietic cancers. Am J Ind Med. 1997. 31:287–95.

4). Preston DL., Kusumi S., Tomonaga M, et al. Cancer incidence in atomic bomb survivors. Part III. Leukemia, lymphoma and multiple myeloma, 1950~ 1987. Radiat Res. 1994. 137:S68–97.
5). Kerr JR., Barah F., Cunniffe VS, et al. Association of acute parvovirus B19 infection with new onset of acute lymphoblastic and myeloblastic leukaemia. J Clin Pathol. 2003. 56:873–5.

6). Quesnel B., Kantarjian H., Bjergaard JP, et al. Therapy-related acute myeloid leukemia with t(8;21), inv (16), and t(8;16): a report on 25 cases and review of the literature. J Clin Oncol. 1993. 11:2370–9.
7). Lilly F., Boyse EA., Old LJ. Genetic basis of susceptibility to viral leukaemogenesis. Lancet. 1964. 2:1207–9.
8). Dorak MT., Lawson T., Machulla HK., Darke C., Mills KI., Burnett AK. Unravelling an HLA-DR association in childhood acute lymphoblastic leukemia. Blood. 1999. 94:694–700.

9). Dorak MT., Chalmers EA., Gaffney D, et al. Human major histocompatibility complex contains several leukemia susceptibility genes. Leuk Lymphoma. 1994. 12:211–22.

10). Dorak MT., Machulla HK., Hentschel M., Mills KI., Langner J., Burnett AK. Influence of the major histocompatibility complex on age at onset of chronic lymphoid leukaemia. Int J Cancer. 1996. 65:134–9.

11). Goulmy E., Schipper R., Pool J, et al. Mismatches for minor histocompatibility antigens between HLA-identical donors and recipients and the development of graft-versus-host disease after bone marrow transplantation. N Engl J. 1996. 334:281–5.
13). Warren EH., Gavin M., Greenberg PD., Riddell SR. Minor histocompatibility antigens as targets for T-cell therapy after bone marrow transplantation. Curr Opin Hematol. 1998. 5:429–33.

14). Di Terlizzi S., Zino E., Mazzi B, et al. Therapeutic and diagnostic applications of minor histocompatibility antigen HA-1 and HA-2 disparities in allogeneic hematopoietic stem cell transplantation: a survey of different populations. Biol Blood Marrow Transplant. 2006. 12:95–101.

15). Pietz BC., Warden MB., DuChateau BK., Ellis TM. Multiplex genotyping of human minor histocompatibility antigens. Hum Immunol. 2005. 66:1174–82.

16). Dolstra H., Fredrix H., Maas F, et al. A human minor histocompatibility antigen specific for B cell acute lymphoblastic leukemia. J Exp Med. 1999. 189:301–8.

17). Dolstra H., de Rijke B., Fredrix H, et al. Bi-directional allelic recognition of the human minor histocompatibility antigen HB-1 by cytotoxic T lymphocytes. Eur J Immunol. 2002. 32:2748–58.

18). Miyazawa M., Nishio J., Chesebro B. Genetic control of T cell responsiveness to the Friend murine leukemia virus envelope antigen. Identification of class II loci of the H-2 as immune response genes. J Exp Med. 1998. 168:1587–605.

19). Chesebro B., Kaiser HE. Neoplasms-comparative pathology of growth in animals, plants, and man. Influence of the major histocompatibility complex (H-2) on oncornavirus-induced neoplasia in mice. Baltimore: Williams and Wilkins. 1981. 475–82.
20). Demant P., Oomen LC., Oudshoorn-Snoek M. Genetics of tumor susceptibility in the mouse: MHC and non-MHC genes. Adv Cancer Res. 1989. 53:117–79.

21). Dorak MT., Oguz FS., Yalman N, et al. A male-specific increase in the HLA-DRB4 (DR53) frequency in high-risk and relapsed childhood ALL. Leuk Res. 2002. 26:651–6.

22). Posthuma EF., Falkenburg JH., Apperley JF, et al. HLA-DR4 is associated with a diminished risk of the development of chronic myeloid leukemia (CML). Chronic Leukemia Working Party of the European Blood and Marrow Transplant Registry. Leukemia. 2000. 14:859–62.
23). den Haan JM., Meadows LM., Wang W, et al. The minor histocompatibility antigen HA-1: a diallelic gene with a single amino acid polymorphism. Science. 1998. 279:1054–7.

Go to : 
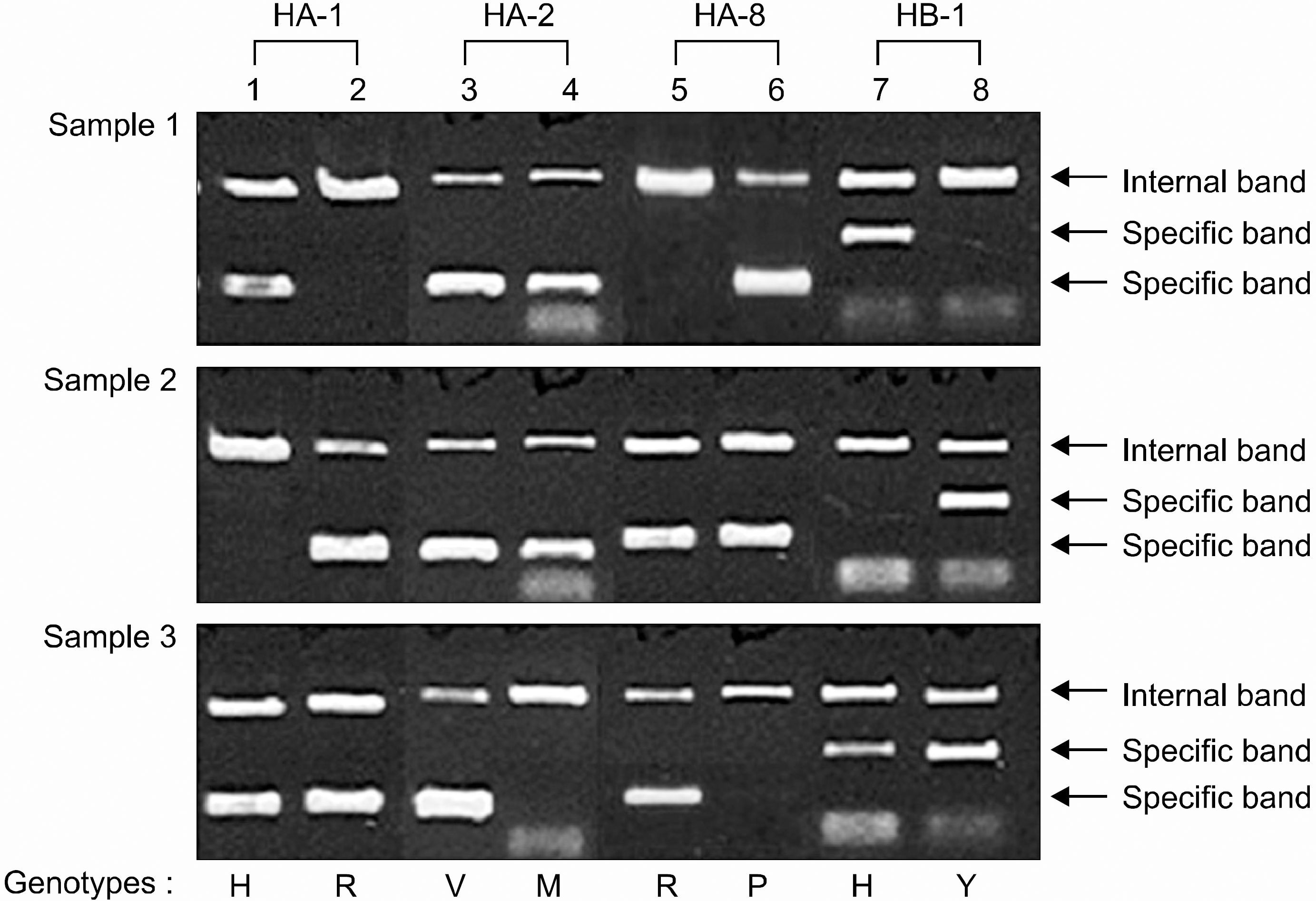 | Fig 1.Genotyping of HA-1, -2, -8 and HB-1 by PCR-SSP. Lane 1: H type for HA-1, lane 2: R type for HA-1, lane 3: V type for HA-2, lane 4: M type for HA-2, lane 5: R type for HA-8, lane 6: P type for HA-8, lane 7: H type for HB-1, lane 8: Y type for HB-1. Each type of mHags HA-1, -2, -8 and HB-1 of the three samples was as follows; sample 1: HH (HA-1), VM (HA-2), PP (HA-8), HH (HB-1); sample 2: RR (HA-1), VM (HA-2), RP (HA-8), YY (HB-1); sample 3: HR (HA-1), VV (HA-2), RR, (HA-8), HY (HB-1). |
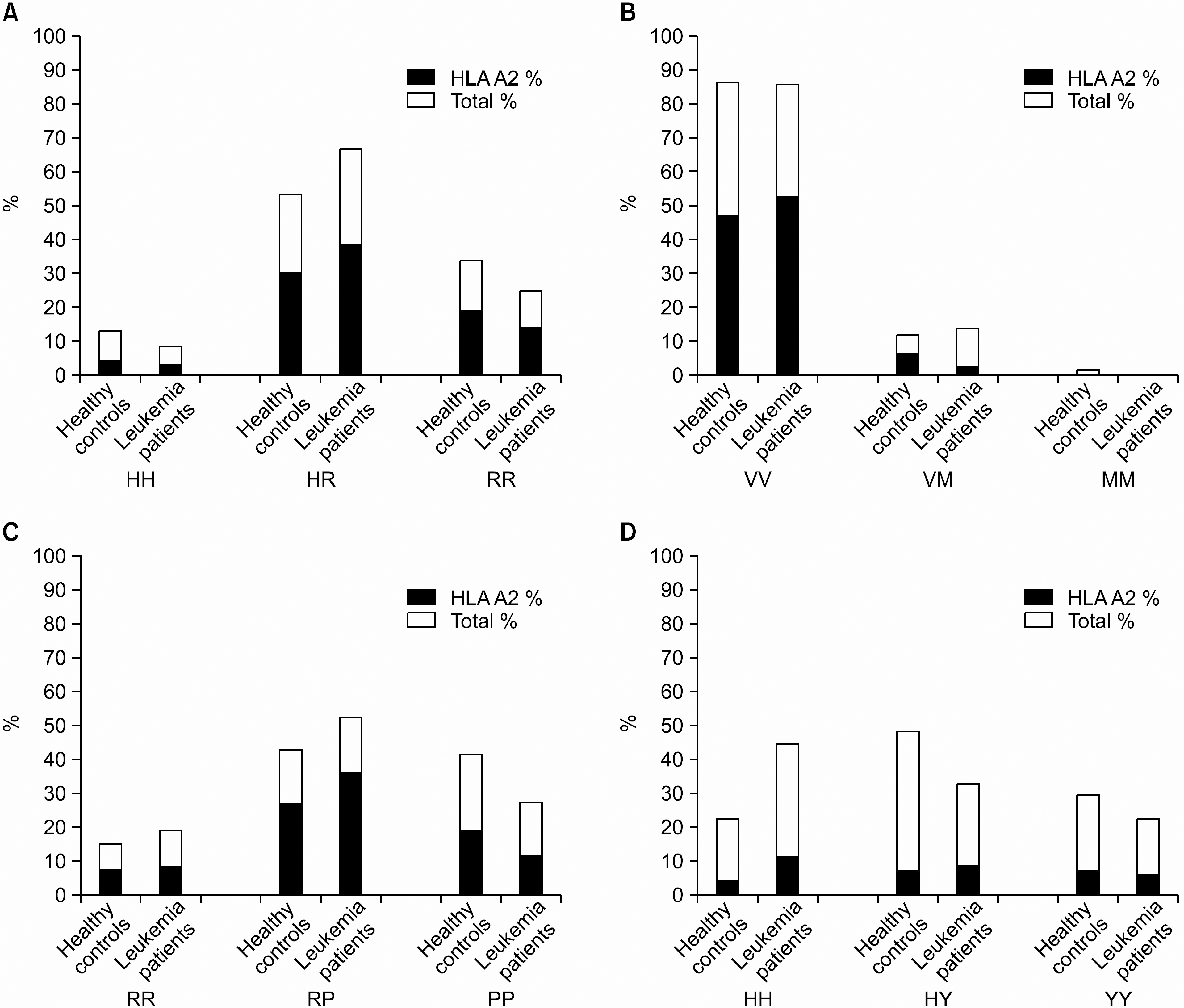 | Fig. 2Allelic frequencies of HA-1, -2, -8 and HB-1 in healthy controls (n=139) and patients with leukemia (n=36) in the Korea. |
Table 1.
Primer sets for genotyping of HA-1, HA-2, HA-8, and HB-1
Table 2.
Genotype and phenotype frequencies of HA-1 in healthy controls and patients with leukemia in Korea
Table 3.
Genotype and phenotype frequencies of HA-2 in healthy controls and patients with leukemia in Korea
Table 4.
Genotype and phenotype frequencies of HA-8 in healthy controls and patients with leukemia in Korea
Table 5.
Genotype and phenotype frequencies of HB-1 in healthy controls and patients with leukemia in Korea
| Genotype frequency n (%) | ||||||
|---|---|---|---|---|---|---|
| Locus | Healthy controls | Leukemia patients | ||||
| (n=139) | Total (n=36) | ALL (n=12) | AML (n=20) | CML (n=4) | ||
| HB-1 | HH | 31 (22.3) | 16 (44.4)∗ | 5 (41.7) | 10 (50)∥ | 1 (25) |
| YH | 67 (48.2) | 12 (33.3) | 2 (16.6) | 7 (35) | 3 (75) | |
| YY | 41 (29.5) | 8 (22.3) | 5 (41.7) | 3 (15) | 0 (0) | |
| H | 98 | 28 | 7 | 17 | 4 | |
| Y | 108 | 20† | 7 | 10¶ | 3 | |
| Gene frequency n (%) | ||||||
| H | 129 (46.4) | 44 (61.1)‡ | 12 (50) | 27 (67.5)∗∗ | 5 (62.5) | |
| Y | 149 (53.6) | 28 (38.9)§ | 12 (50) | 13 (32.5)†† | 3 (37.5) | |
∗P-value was calculated by chi-square tests of total leukemia patients versus healthy controls (P=0.008, OR=2.8, CI=1.32~ 5.91);
† P-value was calculated by chi-square tests of total leukemia patients versus healthy controls (P=0.008, OR=0.36, CI=0.17~0.76);
‡ P-value was calculated by chi-square tests of total leukemia patients versus healthy controls (P=0.03, OR=1.82, CI=1.08~3.06);
§ P-value was calculated by chi-square tests of total leukemia patients versus healthy controls (P=0.03, OR=0.56, CI=0.33~0.93);




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download