Abstract
Background:
The prevalent natural killer (NK) cells induce alloreaction against leukemic cells during post-transplant. NK cell alloreactivity depends on the compatibility of killer cell immunoglobulin-like receptors (KIR) epitopes for graft-versus-host disease. Genotypic expressions of inhibitory or activating KIR in patients with acute myelogenous leukemia (AML) and their HLA-matched sibling donors, as a model for Korean KIR haplotype diversity and NK alloreactivity, were investigated.
Methods:
Ninety-two patients in complete remission and their 76 HLA-matched sibling donors were enrolled in this study. All the patients were scheduled to receive allogeneic hematopoietic stem cell transplantations (HSCT). KIR PCR-SSP typing was performed for 19 different kinds of KIR genes and pseudogenes. The PCR data representing the KIR genotypes from both the patients and donors were compared.
Results:
We found 43 Korean KIR haplotypes. Thirty-three variable haplotypes for the AML patients, in addition to 25 haplotypes for the normal HSCT donors, were demonstrated. Of note, the expressions of specific genes such as 2DL2 (P=0.026), 2DS2 (P=0.042), and 2DS4 (P=0.037) revealed remarkable differences between the patients and the normal donors. Korean HLA-identical sibling pairs showed 38% KIR matches in terms of the gene content and allelic polymorphism. Although the KIR gene content was the same between the patients and the donors, 40% of those matched pairs of patients and donors showed allelic polymorphism, specifically in the context of 2DL5 and 2DS4 genes.
Go to : 
REFERENCES
1). Ruggeri L., Capanni M., Urbani E, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002. 295:2097–100.

2). Parham P., McQeen KL. Alloreactive killer cells: hindrance and help for haematopoietic transplants. Nat Rev Immunol. 2003. 3:108–22.

3). Godfrey DI., MacDonald HR., Kronenberg M., Smyth MJ., Van Kaer L. NKT cells: what's in a name? Nat Rev Immunol. 2004. 4:231–7.

4). Degli-Esposti MA., Smyth MJ. Close encounters of different kinds: dendritic cells and NK cells take centre stage. Nat Rev Immunol. 2005. 5:112–24.

5). Farag SS., Fehniger TA., Ruggeri L., Velardi A., Caligiuri MA. Natural killer cell receptors: new biology and insights into the graft-versus-leukemia effect. Blood. 2002. 100:1935–47.

6). Ljunggren HG., Karre K. In search of the ‘missing self': MHC molecules and NK cell recognition. Immunol Today. 1990. 11:237–44.

7). Gagne K., Brizard G., Gueglio B, et al. Relevance of KIR gene polymorphisms in bone marrow transplantation outcome. Hum Immunol. 2002. 63:271–80.

8). Aversa F., Tabilio A., Velardi A, et al. Treatment of high-risk acute leukemia with T-cell-depleted stem cells from related donors with one fully mismatched HLA haplotype. N Engl J Med. 1998. 339:1186–93.

9). Hsu KC., Chida S., Geraghty DE., Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev. 2002. 190:40–52.

10). Kim HJ., Min WS., Kim YJ., Kim DW., Lee JW., Kim CC. Haplotype mismatched transplantation using high doses of peripheral blood CD34+ cells together with stratified conditioning regimens for high-risk adult acute myeloid leukemia patients: a pilot study in a single Korean institution. Bone Marrow Transplant. 2005. 35:959–64.

Go to : 
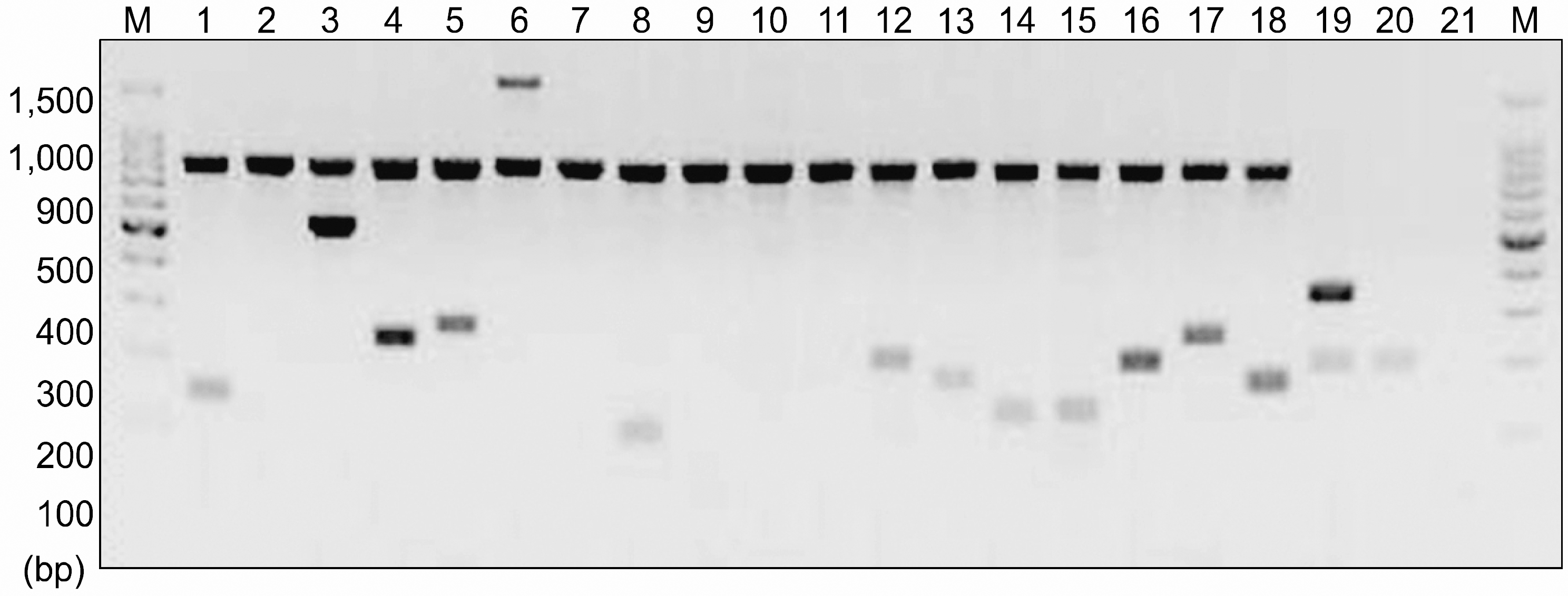 | Fig. 1KIR genotyping by PCR-SSP in a specific AML patient. Details are described in ‘Materials and Methods’. 1, 2DL1; 2, 2DL2; 3, 2DL3; 4, 2DL4; 5, 2DL5A; 6, 2DL5B∗002/004; 7, 2DL5B∗003; 8, 2DS1; 9, 2DS2; 10, 2DS3; 11, 2DS4∗ 00101/00102/002; 12, 2 DS4∗003; 13, 2DS5; 14, 3DL1; 15, 3DL2; 16, 3DL3; 17, 3DS1; 18, 2DP1; 19, 3DP1∗001/001; 20, 3DP1∗00301/00302; 21, negative control; M, marker. |
Table 1.
Summarized data of KIR genotyping from normal transplantation donors in this study (n=76)
Table 2.
Summarized data of KIR genotyping from acute myelogenous leukemia patients in this study (n=92)
Table 3.
Summarized data of KIR genotyping from all enrolled individuals in this study (n=168)




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download