Abstract
Background
Detection of variable number of tandem repeats (VNTR) between recipient and donor has been adopted to monitor the degree of chimerism after allogeneic stem cell transplantation (SCT). In allogeneic SCT, besides MHC-disparity, the disparity of various polymorphous proteins encoded by several genes may play a critical role in the pathogenesis of graft-versus-host disease (GVHD). However, the biologic effect of VNTR disparity has been scarcely studied.
Methods
We analyzed 84 patients receiving SCT from HLA-identical sibling (n=68) or unrelated donors (n=16). Enrolled diseases included AML 48, ALL 8, CML 15, NHL 10, and high-risk MDS 3. The PCR was performed to amplify 3 VNTR regions (D1S80, D1S111, and D17S5).
Results
We observed strong correlation between the D1S80 disparity and transplant outcomes in terms of OS (P=0.0179) or non-relapse mortality (NRM) (P=0.0305), but not for D1S111 or D17S5 disparity. The D1S80-fully matched pair showed a better OS (72% vs 38%) and lower NRM (17% vs 50%) compared to partially matched or mismatched pairs. In multivariate analyses, D1S80-fully matched pair was found to be independent favorable prognostic factor for OS (P=0.03) or NRM (P=0.05). In addition, the D1S80 disparity was significantly associated with the myeloid engraftment speed (P=0.01) or the occurrence of gut chronic GVHD (P=0.05).
Go to : 
REFERENCES
1). Kim TY, Park SH, Kwon EH, Kim KY, Suh JS, Sohn SK. The discrimination power and effectiveness of 3 kinds of LTR primers in the VNTR-PCR for evaluation of the engraftment of allogeneic peripheral blood stem cell transplantation. Korean J Clin Pathol. 2001; 21:527–33.
2). Kim TY, Park SH, Suh JS, Sohn SK. A case of DNA chimerism analysis as a marker of donor lymphocyte infusion. Korean J Hematol. 2001; 36:342–5.
3). Martinelli G, Trabetti E, Zaccaria A, et al. In vitro amplification of hypervariable DNA regions for the evaluation of chimerism after allogeneic BMT. Bone Marrow Transplant. 1993; 12:115–20.
4). van Leeuwen JE, van Tol MJ, Joosten AM, et al. Relationship between patterns of engraftment in peripheral blood and immune reconstitution after allogeneic bone marrow transplantation for (severe) combined immunodeficiency. Blood. 1994; 84:3936–47.

5). Sreenan JJ, Pettay JD, Tbakhi A, et al. The use of amplified variable number of tandem repeats (VNTR) in the detection of chimerism following bone marrow transplantation. A comparison with restriction fragment length polymorphism (RFLP) by Southern blotting. Am J Clin Pathol. 1997; 107:292–8.

6). Rothberg PG, Gamis AS, Baker D. Use of DNA polymorphisms to monitor engraftment after allogeneic bone marrow transplantation. Clin Lab Med. 1997; 17:109–18.

7). Thiede C. Diagnostic chimerism analysis after allogeneic stem cell transplantation: new methods and markers. Am J Pharmacogenomics. 2004; 4:177–87.
8). Falkenburg JH, van de Corput L, Marijt EW, Willemze R. Minor histocompatibility antigens in human stem cell transplantation. Exp Hematol. 2003; 31:743–51.

9). Chao NJ. Minors come of age: minor histocompatibility antigens and graft-versus-host disease. Biol Blood Marrow Transplant. 2004; 10:215–23.

10). Alcoceba M, Diez-Campelo M, Martin-Jimenez P, et al. Allogeneic transplantation with identical MHC: clinic-pronostic value of discrepances of microsa-tellite DNA regions between recipient and donor. Blood. 2004; 104:912a.

11). Stern M, Meyer-Monard S, Bucher C, et al. Prognostic value of discrepancies in micro-satellite DNA regions between donor and recipient in allogeneic stem cell transplantation. Bone Marrow Transplant. 2005; 35(Suppl 2):S44.
12). Sohn SK, Kim DH, Kim JG, et al. Transplantation outcome in allogeneic PBSCT patients according to a new chronic GVHD grading system, including extensive skin involvement, thrombocytopenia, and progressive-type onset. Bone Marrow Transplant. 2004; 34:63–8.

13). Kim DH, Kim JG, Sohn SK, et al. Clinical impact of early absolute lymphocyte count after allogeneic stem cell transplantation. Br J Haematol. 2004; 125:217–24.

14). Sohn SK, Kim JG, Chae YS, et al. Large-volume leukapheresis using femoral venous access for harvesting peripheral blood stem cells with the Fenwal CS 3000 Plus from normal healthy donors: predictors of CD34+ cell yield and collection efficiency. J Clin Apher. 2003; 18:10–5.

15). Sohn SK, Kim JG, Sung WJ, et al. Harvesting peripheral blood stem cells from healthy donors on 4th day of cytokine mobilization. J Clin Apher. 2003; 18:186–9.

16). Przepiorka D, Weisdorf D, Martin P, et al. 1994 Consensus Conference on Acute GVHD Grading. Bone Marrow Transplant. 1995; 15:825–8.
17). Shulman HM, Sullivan KM, Weiden PL, et al. Chronic graft-versus-host syndrome in man. A longterm clinicopathologic study of 20 Seattle patients. Am J Med. 1980; 69:204–17.
Go to : 
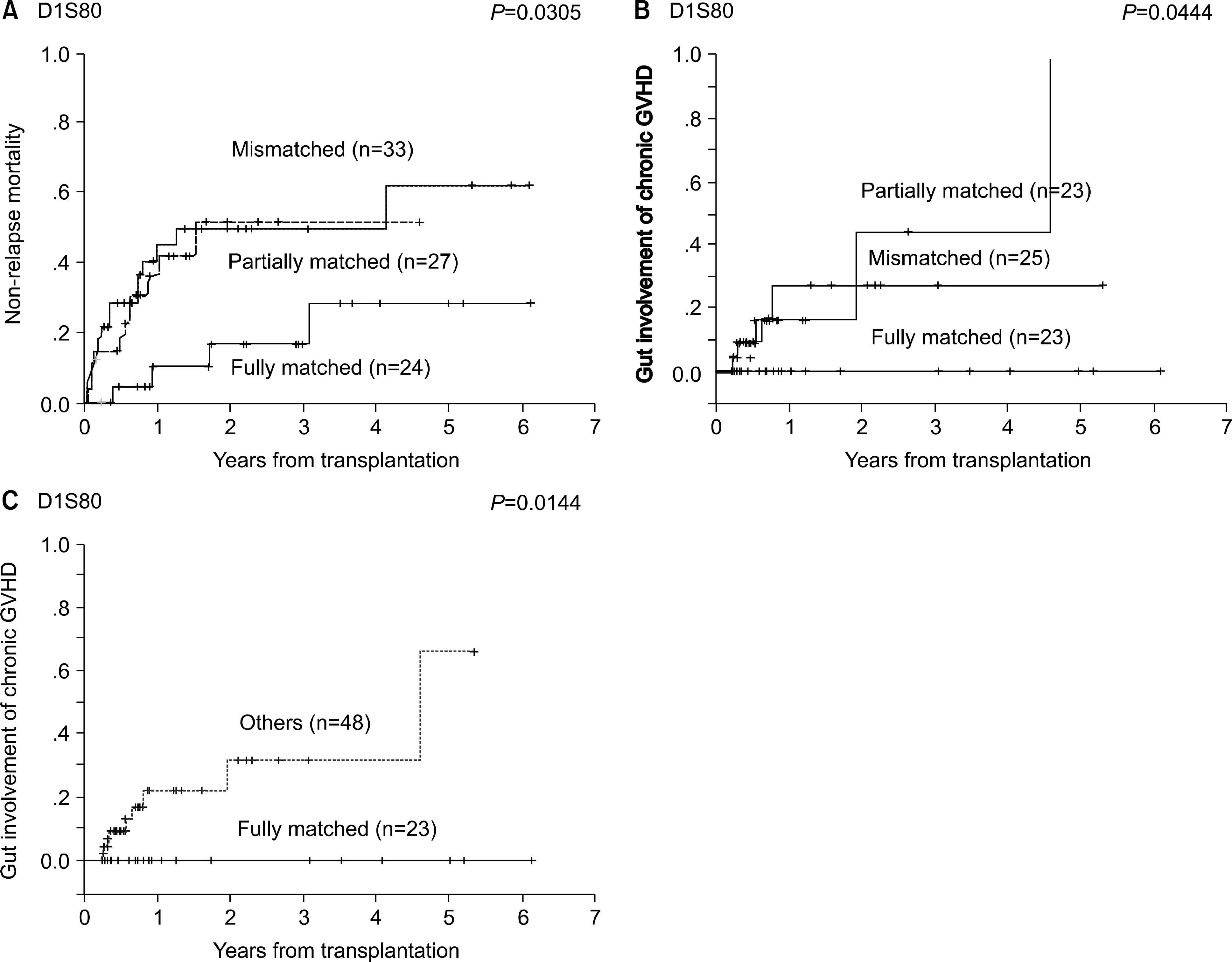 | Fig. 1.The cumulative incidence of non-relapse mortality (A) and occurrence of gut-involved chronic GVHD (B,C) according to the D1S80 disparity. |
Table 1.
Characteristics of the human long tandem repeat markers
Table 2.
The transplant outcomes in terms of overall survival, non-relapse mortality or relapse according to the VNTR disparities
Table 3.
Transplantation outcomes according to VNTR disparity for D1S80 loci
| Disparity of D1S80 | Fully matched pair (n=24, 29%) | Partially matched pair (n=27, 32%) | Mismatched pair (n=33, 39%) | P-value |
|---|---|---|---|---|
| Follow-up duration (days) | 347 (15~2181) | 232 (17~1290) | 479 (15~2181) | |
| Engraftment | ||||
| Myeloid | 13.0 (10~24) | 13.5 (10~25) | 16 (10~30) | 0.01 |
| Platelet | 14 (9~161) | 14 (10~56) | 17 (0~42) | 0.26 |
| Acute GVHD (n=81) | (24) | (26) | (31) | |
| Overall | 21 (88) | 23 (89) | 22 (71) | 0.19 |
| ≥grade 2 | 17 (71) | 20 (77) | 20 (65) | 0.59 |
| ≥grade 3 | 4 (17) | 7 (27) | 8 (26) | 0.64 |
| Skin,≥stage 2 | 12 (50) | 14 (54) | 15 (48) | 0.92 |
| Liver,≥stage 1 | 4 (17) | 9 (31) | 13 (42) | 0.14 |
| Gut,≥stage 1 | 14 (58) | 16 (62) | 17 (55) | 0.88 |
| Chronic GVHD (n=71) | (23) | (23) | (25) | |
| Limited+extensive | 16 (70) | 19 (83) | 20 (80) | 0.60 |
| Extensive | 9 (39) | 11 (48) | 11 (44) | 0.87 |
| Skin involvement | 11 (48) | 13 (57) | 11 (44) | 0.68 |
| Hepatic involvement | 11 (48) | 12 (52) | 14 (56) | 0.85 |
| Gut involvement | 0 (0) | 5 (22) | 4 (16) | 0.05 |
| Infectious events | ||||
| CMV reactivation | 15 (63) | 15 (56) | 17 (52) | 0.71 |
| Infectious events | 11 (46) | 10 (37) | 18 (55) | 0.40 |
| Bacterial infections | 6 (25) | 7 (26) | 10 (30) | 0.91 |
| Viral infections | 6 (25) | 3 (11) | 10 (30) | 0.21 |
| Fungal infections | 3 (13) | 3 (11) | 4 (12) | 1.00 |
| Survival | ||||
| Relapse | 8 (33) | 10 (26) | 6 (29) | 0.84 |
| Deaths | 7 (29) | 15 (56) | 21 (64) | 0.03 |
| Causes of death | ||||
| Non-relapse mortalities | 4 (17) | 11 (41) | 15 (45) | 0.06‡ |
| GVHD+/-infection | 4 (17) | 10 (37) | 13 (39) | 0.15§ |
| Others | 0 (0) | 1 (4)∗ | 2 (6)† | 0.78 |
| Progression | 3 (12) | 4 (15) | 6 (18) | 0.93 |
Table 4.
Multivariate survival analysis of the prognostic factor for overall survival and the cumulative incidence of non-relapse mortality or relapse in overall patients
For the analysis, the D1S80 disparity (fully matched pair vs partially matched or mismatched pairs), the disease status (standard vs advanced risk), transplanted dose of CD34+ cells (6×106/kg), the donor type (sibling vs unrelated donors) and GVHD (acute GVHD grade 0~2 vs 3,4, and the development of chronic GVHD) were included.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download