Abstract
Background
Biomarkers of lymphomas identified by immunostaining of lymphoma tissues were recently found to have a prognostic value for diffuse large B-cell lymphoma (DLBL). Thus, it seems likely that the prognostic prediction of lymphomas might be improved by incorporating biological markers into well known prognostic systems.
Methods
To determine the clinical significance of the biological markers expressed in DLBL, 26 patients, with de novo DLBL, were retrospectively studied at the Chungnam National University Hospital. Archival specimens from the patients were stained with antibodies for the bcl-2, bcl-6, Ki-67, CD 10, IRF-4, Granzyme-B, MHC-II and p16 antigens. Two immunophenotypic patterns of DLBL were identified by the pattern of differentiation; the germinal center (GC, CD10±/Bcl-6+/IRF-4-)-like subgroup and the post germinal center (pGC, CD10±/bcl-6±/IRF4+)-like subgroup.
Results
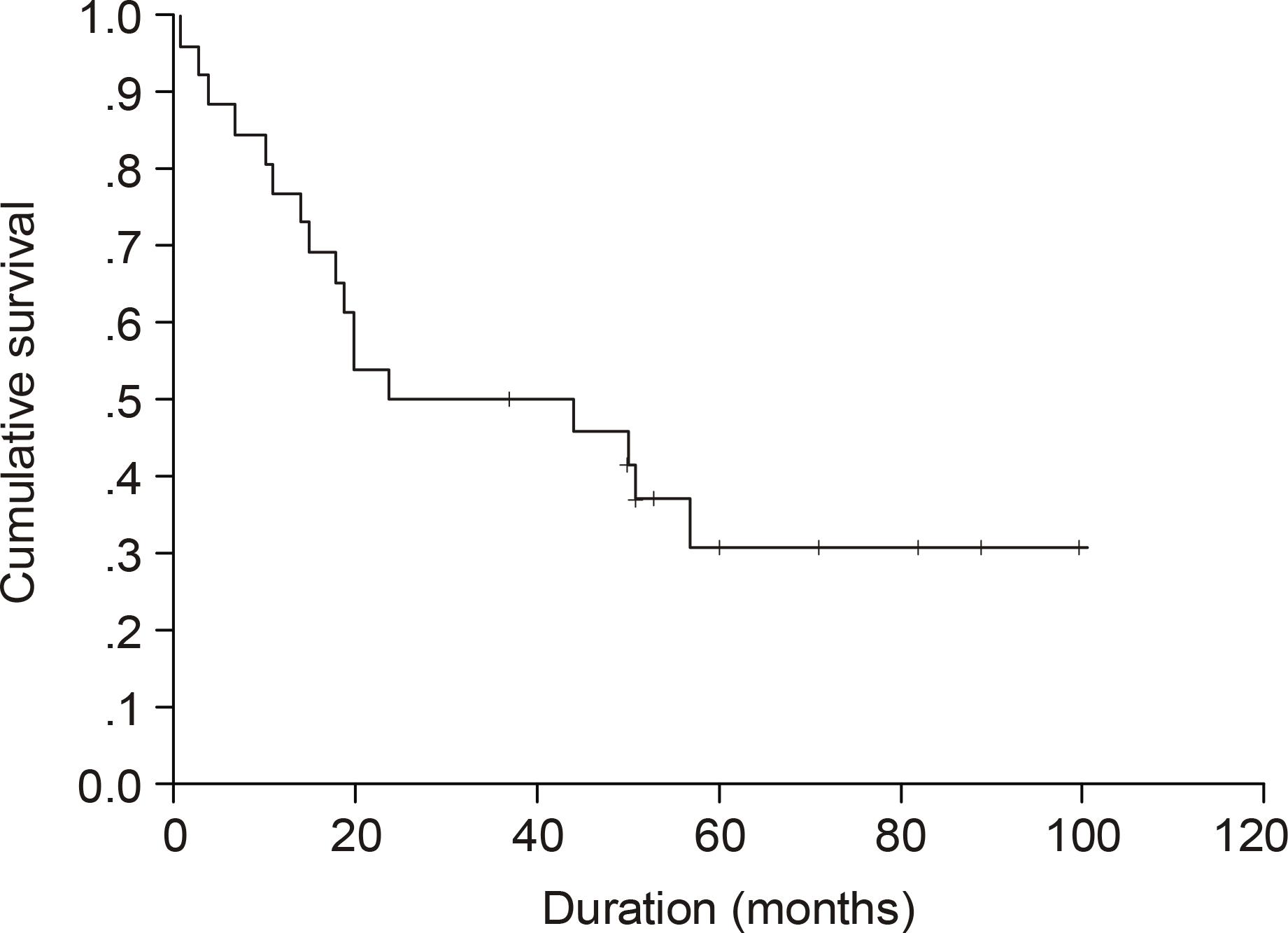
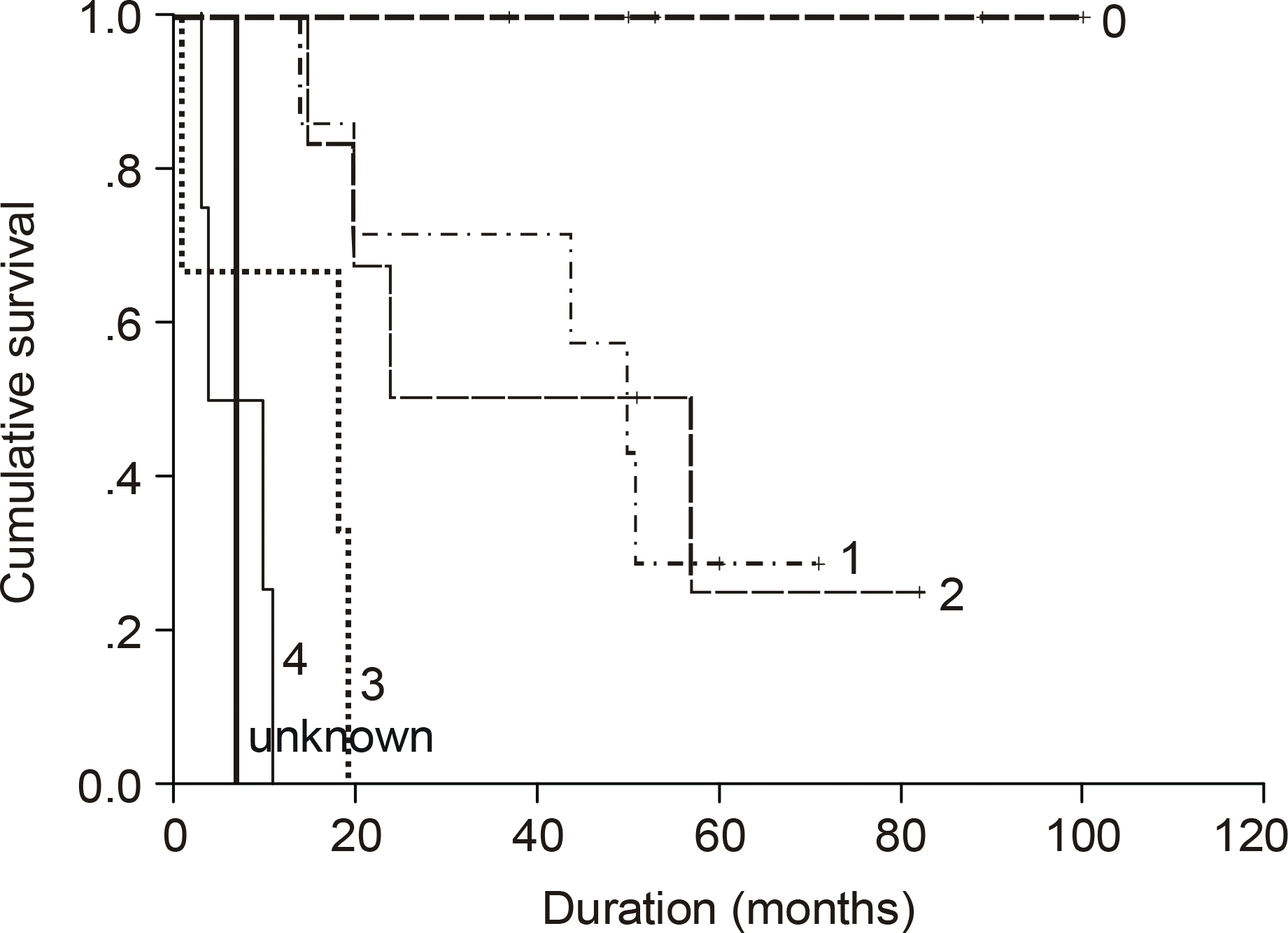
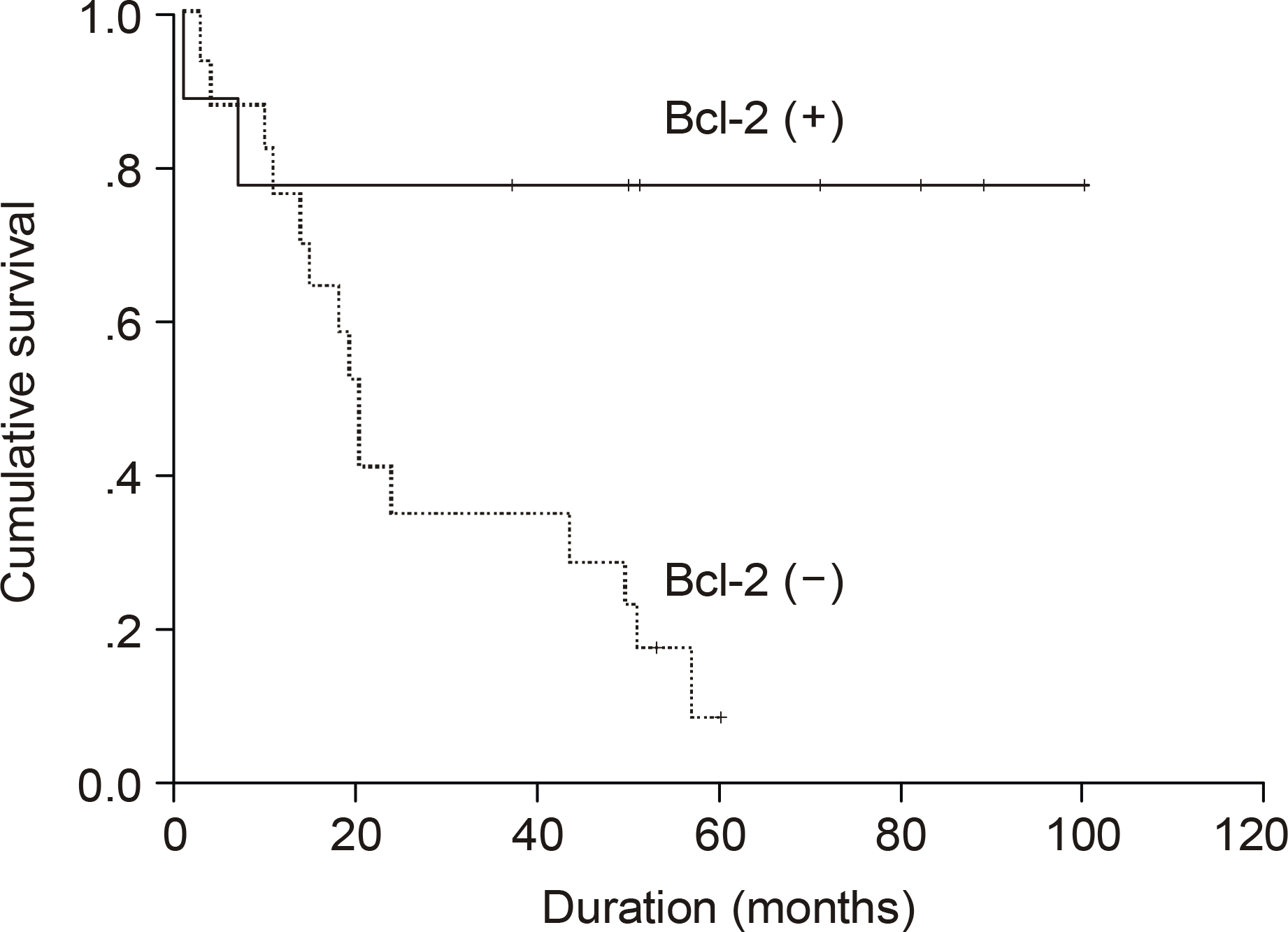
The median age of the subjects was 56 years, ranging form 37 to 69. After a median follow up duration of 48 months, the median survival time was 44 month, ranging from 1~100 months. The five-year overall survival rate Using the Kaplan-Meier method was 32%. The only biomarker affecting the survival was bcl-2 (P=0.009). The survival of the GC-like subgroup was superior to that of the pGC-like subgroup, but without statistical significance (P=0.064). Among 18 patients with IPI scores 0~ 2, those expressing bcl-2 (P=0.002) and the pGC-like subgroup had a worse prognosis compared to the GC-like subgroup (P=0.049).
Go to : 
REFERENCES
1). The Non-Hodgkin's Lymphoma Classification Project: A clinical evaluation of the International Lymphoma Study Group classification of non-Hodgkin's lymphoma. Blood. 1997; 89:3909–18.
2). Hahn JS, Ko YW, Min YH, et al. Statistical analysis of malignant lymphoma in Korea. Korean J Hematol. 1995; 30:197–214.
3). Pasqualucci L, Neumeister P, Goossens T, et al. Hypermutation of multiple protooncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001; 412:341–6.

4). Harris NL, Jaffe ES, Stein H, et al. A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood. 1994; 84:1361–92.
5). Gatter KC, Warnke RA. Diffuse large B-cell lymphoma. In: Jaffe ES, Harris NL, Stein H, Vardiman JW, eds. WHO Classification of tumors. Pathology and genetics of tumors of haematopoietic and lymphoid tissues. Lyon, France: International Agency for Research on Cancer (IARC) Press. 2006. 171–4.
6). The International Non-Hodgkin's Lymphoma Prognostic Factors Project: a predictive model for aggressive non-Hodgkin's lymphoma. N Engl J Med. 1993; 329:987–94.
7). Fisher RI, Gaynor ER, Dahlberg S, et al. Comparison of a standard regimen (CHOP) with three intensive chemotherapy regimens for advanced non Hodgkin's lymphoma. N Engl J Med. 1993; 328:1002–6.
8). Fisher RI, Miller TP, O'Connor OA. Diffuse aggressive lymphoma. Hematology Am Soc Hematil Educ Program. 2006. 221–36.

9). Lossos IS, Jones CD, Warnke R, et al. Expression of a single gene, bcl-6, strongly predicts survival in patient with diffuse large B cell lymphoma. Blood. 2001; 98:945–51.
10). Xu Y, McKenna RW, Kroft SH. Comparison of multiparameter flow cytometry with cluster analysis and immunohistochemistry for the detection of CD 10 in diffuse large B-Cell lymphomas. Mod Pathol. 2002; 15:413–9.
11). Mittrucker HW, Matsuyama T, Grossman A, et al. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997; 275:540–3.

12). Falini B, Fizzotti M, Pucciarini A, et al. A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood. 1999; 95:2084–92.

13). Tsujimoto Y, Gorham J, Cossman J, Jaffe E, Crose CM. The t(14;18) chromosome translocation involved in B-cell neoplasms result from mistakes in VDJ joining. Science. 1999; 229:1390–3.
14). Korsmeyer SJ. Bcl-2 initiates a new category of onco-genes: regulators of cell death. Blood. 1992; 80:879–86.

15). Miyashita T, Reed JC. Bcl-2 oncoprotein blocks chemotherapy-induced apoptosis in a human leukemia cell line. Blood. 1993; 81:151–7.

16). Hermine O, Haioun C, Lepage E, et al. Prognostic significance of bcl-2 protein expression in aggressive non-Hodgkin's lymphoma. Blood. 1996; 87:265–72.
17). Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993; 366:704–7.

18). Villuendas R, Sanchez-Beato M, Martinez JC, et al. Loss of p16/INK4A protein expression in non-Hodgkin's lymphomas is a frequent finding associated with tumor progression. Am J Pathol. 1993; 153:887–97.

19). Bladergroen BA, Meijer CJ, ten Berge RL, et al. Expression of the granzyme B inhibitor, protease inhibitor 9, by tumor cells in patients with non-Hodgkin and Hodgkin lymphoma: novel protective mechanism for tumor cells to circumvent the immune system? Blood. 2002; 99:232–7.
20). Momburg F, Herrmann B, Moldenhauer G, Moller P. B-cell lymphomas of high grade malignancy frequently lack HLA-DR, -DP and -DQ antigens and associated in variant chain. Int J Cancer. 1987; 40:598–603.
21). Grogan TM, Lippman SM, Spier CM, et al. Independent prognostic significance of a nuclear proliferation antigen in diffuse large cell lymphomas as determined by the monoclonal antibody Ki-67. Blood. 1987; 71:1157–60.

22). Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000; 403:503–11.

23). Rosenwald A, Wright G, Chan WC, et al. Lymphoma/Leukemia Molecular Profiling Project: the use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002; 346:1937–47.
Go to : 
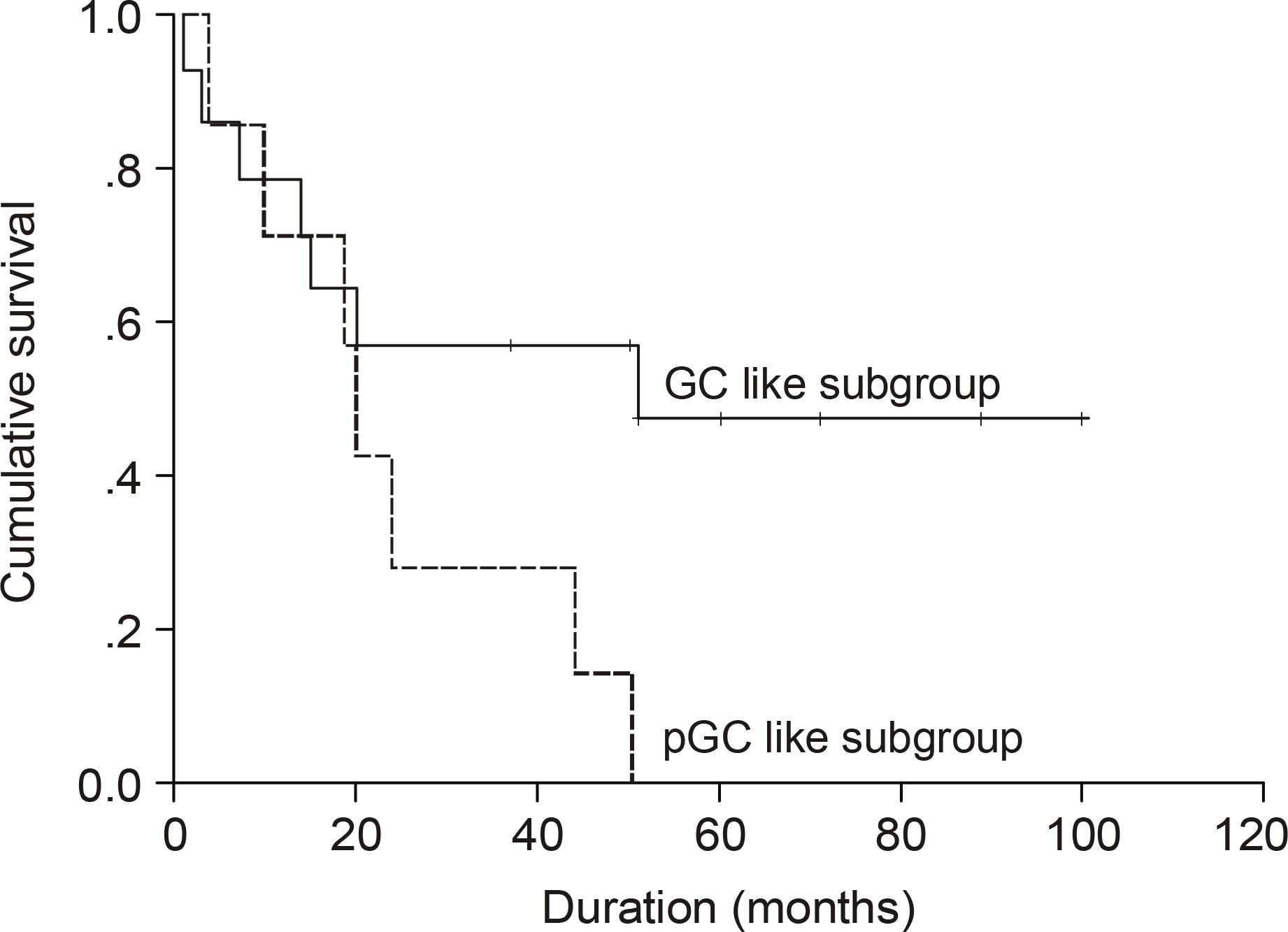 | Fig. 4.Kaplan Meier survival curve according to GC-like subgroup and pGC-like subgroup (P=0.064). The undifferentiated subgroup was excluded. |
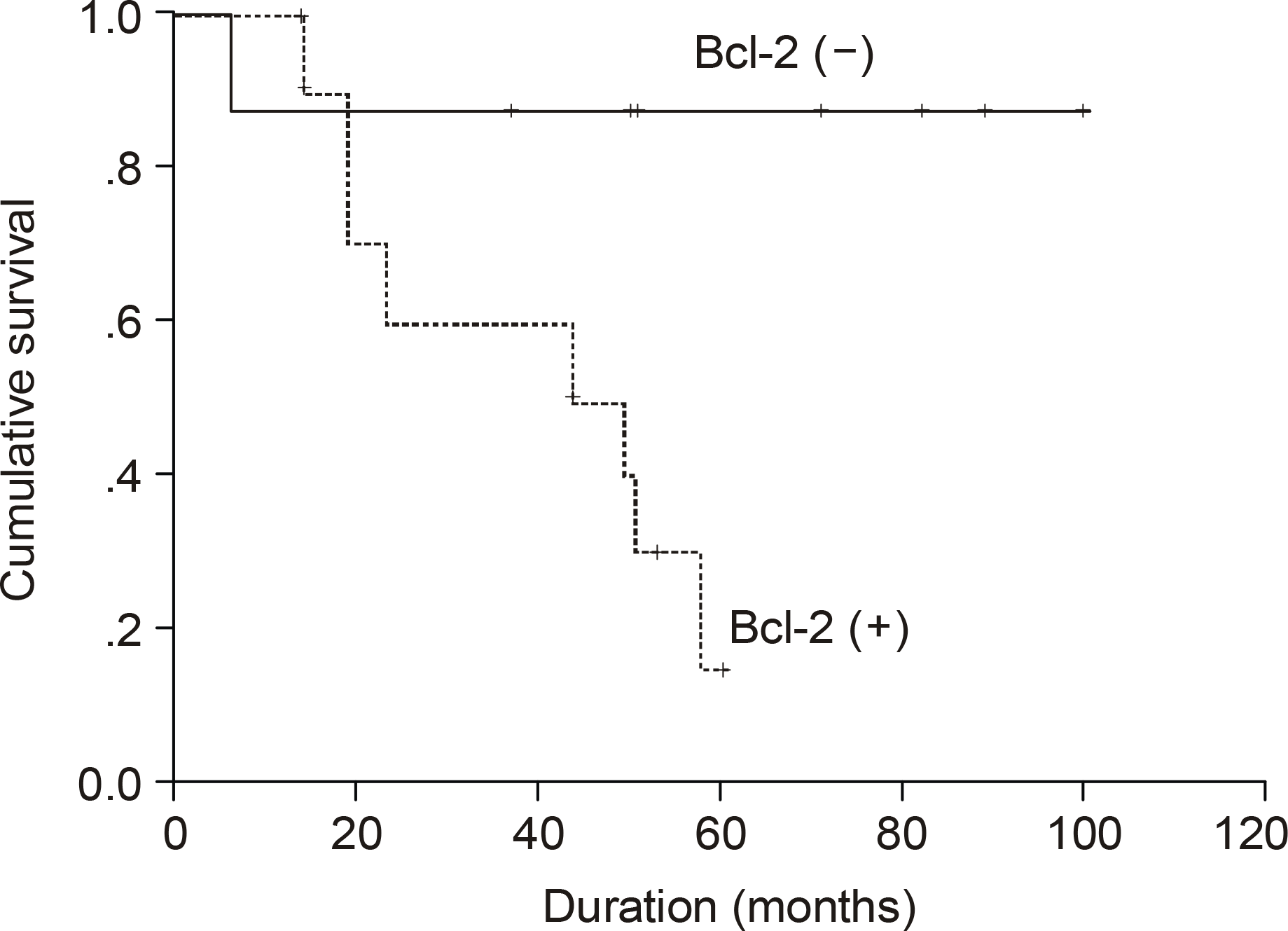 | Fig. 5.Kaplan Meier survival curve according to bcl-2 in patients with IPI score 0~2 (P=0.002). |
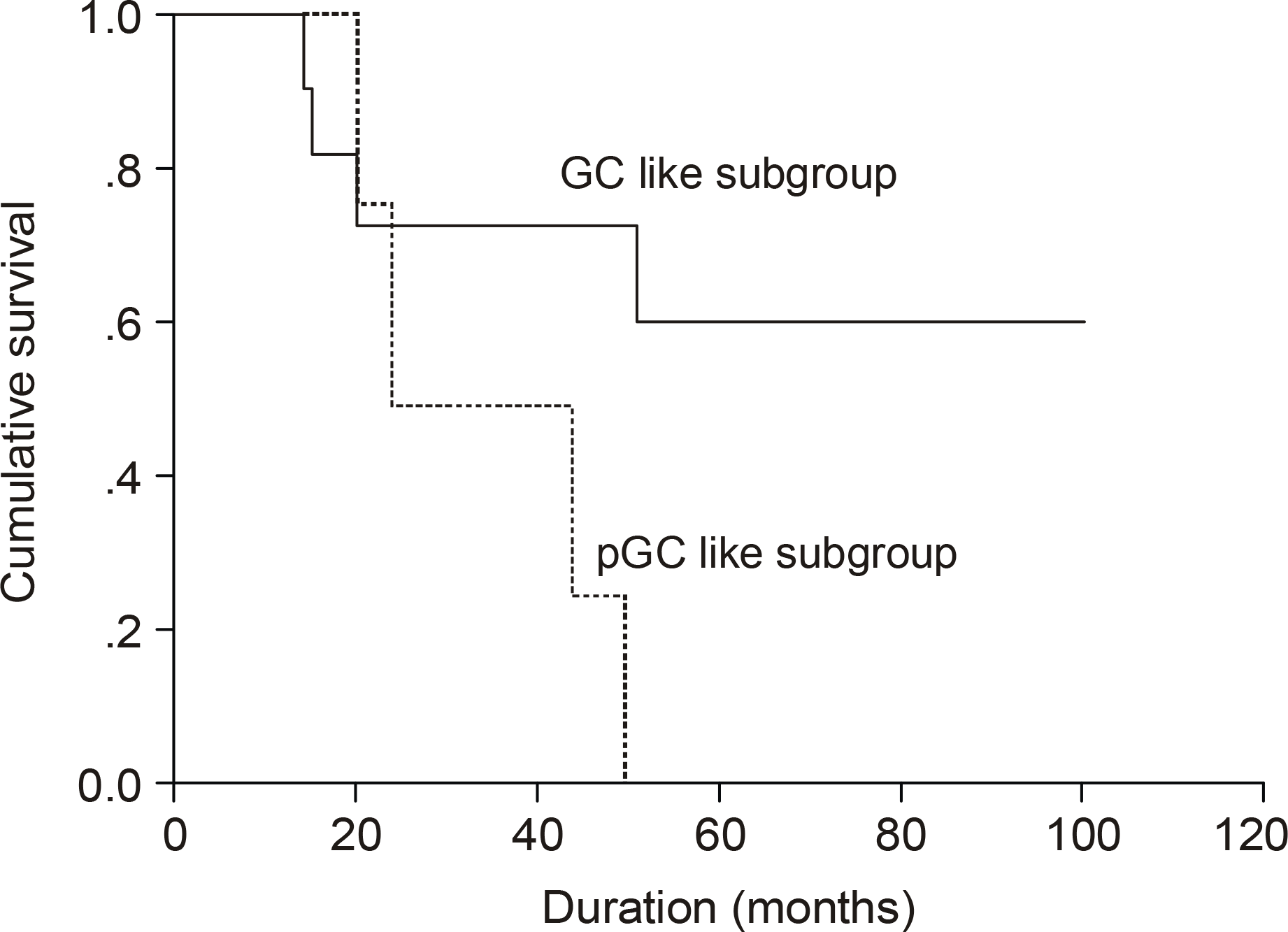 | Fig. 6.Kaplan Meier survival curve according to GC-like subgroup or pGC-like subgroup in patients with IPI score 0~ 2 (P=0.049). The undifferentiated subgroup was excluded. |
Table 1.
The clinical characteristics and factors affecting survival in DLBL patients (No.=26)
Table 2.
The biologic characteristics and markers affecting survival in DLBL patients (No.=26)
| Characteristics | No. of patients (%) | Median survival (month) | P value |
|---|---|---|---|
| Bcl-2 | 0.009 | ||
| Positive | 17 (65.4) | 20 | |
| Negative | 9 (34.6) | Not reached | |
| Bcl-6 | 0.425 | ||
| Positive | 20 (76.9) | 24 | |
| Negative | 6 (23.1) | 18 | |
| Ki-67 | 0.859 | ||
| <60 | 9 (34.6) | 44 | |
| ≥60 | 17 (65.4) | 20 | |
| CD10 | 0.552 | ||
| Positive | 5 (19.2) | Not reached | |
| Negative | 21 (80.8) | 24 | |
| IRF-4 | 0.184 | ||
| Positive | 8 (30.8) | 20 | |
| Negative | 18 (69.2) | 51 | |
| Granzyme-B | 0.533 | ||
| 1+ | 17 (65.4) | 50 | |
| 2+ | 5 (19.2) | 18 | |
| 3+ | 4 (15.4) | 11 | |
| MHC-II | 0.447 | ||
| Positive | 23 (88.5) | 44 | |
| Negative | 3 (11.5) | 10 | |
| p16 | 0.372 | ||
| Positive | 4 (15.4) | 44 | |
| Negative | 22 (84.6) | 20 | |
| Subgroup | 0.064∗ | ||
| GC group | 14 (53.8) | 51 | |
| pGC group | 7 (27.0) | 20 | |
| Undifferentiated | 5 (19.2) |
Table 3.
The biologic characteristics and markers affecting survival in DLBL patients with IPI score 0~2 (No.=18)
| Characteristics | No. of patients (%) | Median survival (month) | P value |
|---|---|---|---|
| Bcl-2 | 0.002 | ||
| Positive | 11 (61.1) | 44 | |
| Negative | 7 (38.9) | Not reached | |
| Bcl-6 | 0.370 | ||
| Positive | 15 (83.3) | 51 | |
| Negative | 3 (16.7) | Not reached | |
| Ki-67 | 0.352 | ||
| <60 | 7 (38.9) | 51 | |
| ≥60 | 11 (61.1) | Not reached | |
| CD 10 | 0.169 | ||
| Positive | 3 (16.7) | Not reached | |
| Negative | 15 (83.3) | 51 | |
| IRF-4 | 0.101 | ||
| Positive | 5 (27.8) | 44 | |
| Negative | 13 (72.2) | Not reached | |
| Granzyme B | 0.381 | ||
| 1+ | 14 (77.8) | 51 | |
| 2+ | 2 (11.1) | 15 | |
| 3+ | 2 (11.1) | Not reached | |
| MHC-II | 0.418 | ||
| Positive | 17 (94.4) | 57 | |
| Negative | 1 (5.6) | Not reached | |
| p16 | 0.448 | ||
| Positive | 3 (16.7) | Not reached | |
| Negative | 15 (83.3) | 57 | |
| Subgroup∗ | 0.049 | ||
| GC | 11 (61.1) | Not reached | |
| pGC | 4 (22.2) | 24 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download