Abstract
Purpose
Hepatitis C virus (HCV) infection is a major cause of liver disease. Several miRNAs have been found to be associated with HCV infection. This study aimed to investigate the functional roles and possible molecular mechanisms of miR-215 in HCV replication.
Materials and Methods
The expression levels of miR-215 and TRIM22 were detected by quantitative real-time PCR (qRT-PCR) and western blot analysis in Con1b subgenomic genotype 1b HCV replicon cells (Con1b cells) and JFH1 full genome infecting Huh7.5.1 cells (Huh7.5.1 cells). HCV RNA levels were measured by qRT-PCR. The protein levels of NS3, NS5A, p65 subunit of NF-κB (p65), and phosphorylated p65 (p-p65) were determined by western blot analysis. The relationship between miR-215 and TRIM22 were explored by target prediction and luciferase reporter analysis.
Results
miR-215 overexpression enhanced HCV replication in Con1b cells, while miR-215 knockdown suppressed HCV replication in Huh7.5.1 cells. TRIM22 was confirmed to be a direct target of miR-215. TRIM22 upregulation resulted in a decline in HCV replication, while TRIM22 inhibition led to enhancement of HCV replication. Additionally, exogenous expression of TRIM22 reversed the facilitating effect of miR-215 on HCV replication, while TRIM22 downregulation counteracted the inhibitory effect of miR-215 knockdown on HCV replication. Furthermore, miR-215 targeted TRIM22 to block the NF-κB pathway, and exerted a positively regulatory role on HCV replication.
Hepatitis C virus (HCV), a hepatotropic RNA virus of the genus Hepacivirus in the Flaviviridae family, is a serious global health problem, with approximately 170–200 million individuals infected worldwide.1 HCV infection has been reported to be able to trigger acute and chronic hepatitis, liver cirrhosis, and even hepatocellular carcinoma.2 Despite enormous advances in diagnosis and therapy, there are still many obstacles to the clinical application of antivirals.34 Therefore, understanding the molecular pathogenesis and identifying effective molecular targets for HCV treatment are of great significance.
microRNAs (miRNAs), a class of endogenous short RNAs consisting of 18–25 nucleotides without protein-coding capacity, downregulate gene expression through translation inhibition or mRNA degradation by binding to the 3'-untranslated region (3'UTR) of target mRNA.5 Increasing evidence suggests that miRNAs play essential roles in a variety of physiological and pathological processes, ranging from metabolic disorders to viral infections.6 Recently, miRNAs have been shown to be associated with HCV infection and host-cell fate:7 For example, miR-122 facilitated HCV replication by binding to the 5'non-coding region of the viral genome.8 A previous study demonstrated that expressions of miR-149 and miR-638 were advantageous for HCV propagation, while miR-24 repressed HCV replication.9 Additionally, miR-130a inhibited HCV replication in both HCV replicon and J6-/JFH1-infected cells by restoring host innate immune responses.10 miR-215, identified at chromosome 1q41, has been proven as a tumor suppressor in epithelial ovarian cancer,11 breast cancer,12 and colorectal cancer13 and as an oncogene in gastric cancer14 and high-grade glioma.15 Moreover, Ishida, et al.16 demonstrated that miR-215 is upregulated in HCV-infected Huh7 cells using miR-NA array analysis and found that miR-215 could increase HCV replication. However, the functions and mechanisms of miR-215 in HCV replication require further investigation.
The tripartite motif (TRIM) protein family is defined by a highly conserved RBCC signature domain, containing a RING finger domain, one or two B-Box domains, and a coiled-coil domain.17 TRIM proteins are implicated in several diseases wherein they have been shown to modulate intracellular events, such as signal transduction, protein quality control, transcription, cell cycle, apoptosis, and development.18 In addition, several TRIM proteins have been found to exhibit anti-viral activity, and may be involved in innate immunity response.19 TRIM22 was previously identified as a natural antiviral effector of human immunodeficiency virus (HIV) and hepatitis B virus (HBV).2021 Moreover, Yang and his colleagues22 uncovered that TRIM22 upregulation suppressed HCV replication possibly by ubiquitination of NS5A. Nevertheless, the molecular mechanisms thereof are still unknown.
In the present study, we aimed to explore the functional roles of miR-215 in HCV replication and its underlying molecular mechanisms. Our study demonstrated that miR-215 promotes HCV replication by targeting TRIM22, providing a potential therapeutic target for HCV infection.
Con1b subgenomic genotype 1b HCV replicon cell line (Con1b) containing the full-length HCV genotype 1b replicon was obtained from Dr. Ian McGilvray (University of Toronto, Canada). The JFH1 full genome infecting Huh7.5.1 cells were generated as previously described.23 Con1b and Huh7.5.1 cells were cultured in DMEM (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum, penicillin (100 U/mL), and streptomycin (100 μg/mL) at 37°C in a humidified incubator with 5% CO2.
To upregulate or suppress miR-215 expression, miR-215 mimics (miR-215) or miR-215 inhibitor (anti-miR-215) was respectively transfected into Con1b and Huh7.5.1 cells, with scrambled miRNAs (miR-con or anti-miR-con) as a corresponding control. To overexpress TRIM22, pcDNA3.1-TRIM22 (TRIM22) was introduced into Con1b and Huh7.5.1 cells, with pcDNA3.1 empty vector (Vector) as a control. To downregulate TRIM22 expression, siRNA specific targeting TRIM22 (si-TRIM22) was transfected into Con1b and Huh7.5.1 cells, with non-specific scrambled siRNA (si-con) as a control. All plasmids and oligonucleotides (Genechem, Inc., Shanghai, China) were transfected into cells at the indicated concentrations using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol.
Total RNA was isolated from Con1b and Huh7.5.1 cells using TRIzol Reagent (Invitrogen). The cDNAs for mRNA and miR215 were synthesized using random primers (Life Technologies Inc., Rockville, MD, USA) and miRNA-specific stemloop primers (Applied Biosystems, Foster City, CA, USA), respectively. Quantitative real-time PCR (qRT-PCR) reactions for mRNA and miR-215 were performed on the Applied Biosystems 7900 Sequence Detection system (Applied Biosystems) using the SYBR Premix Ex TaqTM kit (TaKaRa, Dalian, China) and TaqMan MicroRNA Assays (Applied Biosystems), respectively. GAPDH and U6 were used as endogenous references to normalize mRNA and miR-215 expression.
Equivalent proteins were separated on a 10% SDS-PAGE and then transferred to nitrocellulose membranes. After incubation with primary and second antibodies, the protein bands were visualized using the ECL detection system (Pierce, Rockford, IL, USA). The primary antibodies were as follows: anti-TRIM22 (HPA003575; Sigma Prestige, St Louis, MO, USA), anti-GAPDH (Clone 6C5; Abcam, Cambridge, MA, USA), antibody against p65 subunit of NF-κB (p65) (SC 372; Santa Cruz Biotechnology, Santa Cruz, CA, USA), and antibody against phosphorylated p65 subunit of NF-κB (p-p65) (#5733; Cell Signaling, Beverly, MA, USA).
TRIM22-3'UTR containing the predicted miR-215 binding sites were amplified and cloned into a pGL3 reporter plasmid. Con1b cells were cotransfected with the wild type (WT) or mutant (MUT) TRIM22-3'UTR reporter vector (WT-TRIM22 or MUT-TRIM22) and miR-215 or miR-con. Huh7.5.1 cells were cotransfected with the WT-TRIM22 or MUT-TRIM22 and anti-miR-215 or anti-miR-con. At 48 h post transfection, luciferase activities were detected using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer's instructions.
All data were derived from at least three independent experiments, and are presented as mean±SD. Student's t test or one-way analysis of variance was used to determine the statistical significance of differences using SPSS software, version 19.0 (IBM Corp., Armonk, NY, USA). A p value <0.05 was considered significant.
To explore the effect of miR-215 on HCV replication, gain- and loss-of-function assays were performed in Con1b and Huh7.5.1 cells. The expression levels of miR-215 were increased in Con1b cells transfected with miR-215 mimics, compared with miR-con (Fig. 1A), while Huh7.5.1 cells introduced with antimiR-215 showed lower miR-215 expression than that of antimiR-con group (Fig. 1B). miR-215 overexpression led to a significant increase in HCV RNA replication, compared with miR-con in Con1b cells (Fig. 1C); however, miR-215 inhibition reduced HCV RNA replication, compared with anti-miR-con in Huh7.5.1 cells (Fig. 1D). To further assess the influence of miR-215 on HCV protein, western blot was performed to detect the expression levels of HCV specific proteins NS3 and NS5A. As expected, overexpression of miR-215 elevated the protein levels of NS3 and NS5A in Con1b cells (Fig. 1E). On the contrary, suppression of miR-215 resulted in a decrease in NS3 and NS5A expression in Huh7.5.1 cells (Fig. 1F). All these data suggested that miR-215 plays a vital role in facilitating HCV replication in Con1b and Huh7.5.1 cells.
It is well known that miRNAs exert their functional roles by suppressing their target genes. Therefore, the miRNA target prediction tool Targetscan was used to search for the potential targets of miR-215. The predicted targets of miR-215 are shown in Table 1. Among these genes, TRIM22 was found to be natural antiviral effectors of HIV-1 and HBV.2021 It was also reported that TRIM22 played an important role in controlling HCV replication.22 Thus, TRIM22 was further explored. The binding sequences of miR-215 in 3'UTR of TRIM22 are displayed in Fig. 2A. To confirm whether TRIM22 was a functional target of miR-215, dual luciferase reporter assay was conducted in Con1b and Huh7.5.1 cells. As expected, miR-215 overexpression significantly restrained the luciferase activity of the wild type reporter (WT-TRIM22) in Con1b cells (Fig. 2B), while the luciferase activity of WT-TRIM22 was enhanced by antimiR-215 in Huh7.5.1 cells (Fig. 2C). However, no change was observed in the luciferase activity of the mutant reporter (MUTTRIM22) following the alteration of miR-215 expression in both Con1b and Huh7.5.1 cells. Then, the effects of miR-215 on TRIM22 expression were further investigated in Con1b and Huh7.5.1 cells. Enforced expression of miR-215 decreased TRIM22 mRNA and protein levels in Con1b cells (Fig. 2D and F). Inversely, downregulation of miR-215 led to a significant increase in TRIM22 mRNA and protein expressions in Huh7.5.1 cells (Fig. 2E and G). Altogether, TRIM22 was confirmed as a direct target of miR-215 in Con1b and Huh7.5.1 cells.
To demonstrate the functional roles of TRIM22 in HCV replication, Con1b cells were transfected with TRIM22-overexpressing plasmid (pcDNA-TRIM22), and Huh7.5.1 cells were transfected with siRNA against TRIM22 (si-TRIM22). TRIM22 overexpression and knockdown was successfully established in Con1b and Huh7.5.1 cells, presented as enhancement of TRIM expression in pcDNA-TRIM22-transfected Con1b cells (Fig. 3A and B) and reduction of TRIM levels in si-TRIM22-introduced Huh7.5.1 cells (Fig. 3E and F). qRT-PCR analysis revealed that HCV RNA levels were lowered by TRIM22 overexpression in Con1b cells (Fig. 3C) and were elevated by TRIM22 knockdown in Huh7.5.1 cells (Fig. 3G). Moreover, exogenous expression of TRIM22 resulted in a significant decrease in NS3 and NS5A protein levels in Con1b cells (Fig. 3D), while TRIM22 downregulation remarkably increased NS3 and NS5A protein levels in Huh7.5.1 cells (Fig. 3H). Taken together, these results indicated that TRIM22 could suppress HCV replication in Con1b and Huh7.5.1 cells.
To further investigate whether the facilitating effect of miR-215 on HCV replication is mediated by TRIM22, Con1b cells were transfected with miR-215 alone or cotransfected with TRIM22; meanwhile, Huh7.5.1 cells were transfected with anti-miR-215 alone or cotransfected with si-TRIM22. Then, qRT-PCR analysis and western blot were performed to detect HCV RNA levels and NS3 and NS5A protein expressions in Con1b and Huh7.5.1 cells. As expected, TRIM22 overexpression reversed the promoting effect of miR-215 on HCV RNA levels and NS3 and NS5A protein expressions in Con1b cells (Fig. 4A and C). Additionally, anti-miR-215-induced decreases in HCV RNA levels and NS3 and NS5A protein expression were recovered following TRIM22 knockdown in Huh7.5.1 cells (Fig. 4B and D). Collectively, miR-215 was deemed to induce HCV replication by inhibiting TRIM22 expression.
A previous study indicated that TRIM22 is a novel NF-κB activator.24 Therefore, we further explored whether miR-215 and TRIM22 exerted their functional roles in HCV replication through regulating NF-κB signaling. miR-215 overexpression inhibited the activity of NF-κB p65 subunit (p-p65/p65), which could be reversed by TRIM22 upregulation in Con1b cells (Fig. 5A). TRIM22 knockdown eliminated the facilitating effect of anti-miR-215 on the activity of p65 (p-p65/p65) in Huh7.5.1 cells (Fig. 5D). To further confirm the role of NF-κB signaling in miR-215/TRIM22-mediated HCV regulation, the NF-κB agonist TNF-α and its antagonist pyrrolidinedithiocarbamate (PDTC) were used to treat Con1b and Huh7.5.1 cells, respectively. As we might expect, activation of NF-κB signaling pathway by TNF-α suppressed HCV RNA and NS3 and NS5A protein expressions in Con1b cells (Fig. 5B and C); however, the levels of HCV RNA and NS3 and NS5A proteins were obviously increased by PDTC-mediated inactivation of NF-κB signaling in Huh7.5.1 cells (Fig. 5E and F). All these data demonstrated that miR-215 facilitates HCV replication by inactivation of NF-κB signaling via TRIM22.
Increasing evidence has highlighted the importance of cellular miRNAs in regulating HCV pathogenesis due to their versatility.25 However, the mechanisms by which miRNAs modulate HCV replication are still far from being elucidated. miRNAs have been discovered to exert their functions by modulating cellular factors related to viral replication or host innate immune responses.2627 In the present study, we demonstrated that miR-215 promotes HCV replication by targeting TRIM22 and suppressing the NF-κB signaling pathway.
Previous studies have shown that miR-215 functions as an oncogene or tumor suppressor depending on different targets. For example, miR-215 could promote glioma cell proliferation, clone formation, and migration, as well as inhibit apoptosis, by targeting PCDH9.28 In contrast, miR-215 has also been found to inhibit NSCLC cell proliferation, invasion, and migration and to promote apoptosis by suppressing ZEB2.29 Recently, miR-215 was found to be upregulated in serum of patients with chronic Hepatitis C and HCV-associated hepatocellular carcinoma.3031 Moreover, a previous study disclosed that miR-215 could promote HCV replication,16 although little is known regarding its mechanism in facilitating HCV replication. Consistent with these studies, we also found that miR-215 overexpression enhances HCV replication in Con1b and Huh7.5.1 cells, as evidenced by elevated HCV RNA, as well as increased NS3 and NS5A protein levels.
Subsequently, we further explored the possible molecular basis of miR-215 in inducing HCV replication. Bioinformatic analysis and luciferase reporter experiments revealed that TRIM22 is a direct of miR-215. TRIM22 was firstly identified as an IFN-inducible protein that restricts HIV-1 transcription.32 Also, the antiviral effects of TRIM22 have been delineated in HIV-1,33 HBV,21 and influenza A virus.34 In line with the report by Yang, et al,22 the present study demonstrated that enforced expression of TRIM22 represses HCV replication in Con1b and Huh7.5.1 cells, embodied by decreased HCV RNA, as well as lower NS3 and NS5A protein levels. Moreover, miR-215-elicited HCV replication was undermined by the upregulation of TRIM22 in Con1b and Huh7.5.1 cells, indicating that miR-215 promotes HCV replication by inhibiting TRIM22 expression. Our study is the first to validate the targeted relationship between miR-215 and TRIM22 in HCV infection.
NF-κB signaling pathway was initially identified as a host factor supporting HCV subgenomic replicon replication in Huh7 and Huh7.5 cells.3536 Moreover, a previous report documented that NF-κB inactivation leads to enhanced HCV replication and increased expression of core proteins.37 However, whether miR-215 and TRIM22 modulate HCV replication through NF-κB signaling pathway is unclear. TRIM22 was previously highlighted as a positive regulator of NF-κB-mediated transcription.24 In this study, we found that miR-215 overexpression blocked NF-κB signaling, while this effect was abrogated following TRIM22 upregulation. Moreover, TNF-α-induced NF-κB stimulation suppressed HCV replication. Therefore, we speculate that miR-215 may exert its function in HCV replication by inactivating NF-κB signaling via inhibition of TRIM22.
In conclusion, miR-215 was found to facilitate HCV replication in Con1b and Huh7.5.1 cells. Moreover, the promotive effect of miR-215 in HCV replication was shown to be mediated by TRIM22/NF-κB pathway. Our study outlines a novel miR-215-TRIM22-NF-κB pathway regulating HCV replication, and may provide a theoretical basis from which to develop targeted therapies against HCV.
Figures and Tables
 | Fig. 1miR-215 promotes HCV replication in Con1b and Huh7.5.1 cells. Con1b cells transfected with miR-con or miR-215 and Huh7.5.1 cells introduced with anti-miR-con or anti-miR-215 were incubated for 48 h. (A and B) qRT-PCR analysis was performed to detect miR-215 expression. (C and D) The levels of HCV RNA were measured by qRT-PCR. (E and F) The expression levels of HCV NS5A and NS3 proteins were determined by western blot analysis.*p<0.05. HCV, hepatitis C virus; qRT-PCR, quantitative real-time PCR. |
 | Fig. 2miR-215 directly targets TRIM22. Con1b cells were transfected with miR-con or miR-215, and Huh7.5.1 cells were introduced with anti-miR-con or anti-miR-215. (A) WT and MUT binding sites of miR-215 in TRIM22 3'UTR are shown. (B and C) The relative luciferase activities were measured in Con1b and Huh7.5.1 cells transfected with WT-TRIM22-3'UTR or MUT-TRIM22-3'UTR. The mRNA (D and E) and protein (F and G) levels of TRIM22 were evaluated by qRT-PCR and western blot in Con1b and Huh7.5.1 cells. *p<0.05. WT, wild-type; MUT, mutant; 3'UTR, 3'-untranslated region; qRT-PCR, quantitative real-time PCR. |
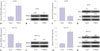 | Fig. 3TRIM22 inhibits HCV replication. Con1b cells were transfected with Vector or TRIM22, followed by detection of TRIM22 mRNA (A) and protein (B), HCV RNA level (C), as well as expressions of NS5A and NS3 proteins (D). Huh7.5.1 cells were introduced with si-con or si-TRIM22, followed by determination of TRIM22 mRNA (E) and protein (F), HCV RNA level (G), as well as expressions of NS5A and NS3 proteins (H). *p<0.05. HCV, hepatitis C virus. |
 | Fig. 4miR-215 promotes HCV replication by targeting TRIM22. (A and B) Con1b cells were transfected with either miR-215 alone or in combination with TRIM22, after which the levels of HCV RNA and NS5A and NS3 proteins were assessed. (C and D) Huh7.5.1 cells were transfected with anti-miR-215 or cotransfected with anti-miR-215 and si-TRIM22, and then the levels of HCV RNA and NS5A and NS3 proteins were measured. *p<0.05. HCV, hepatitis C virus. |
 | Fig. 5miR-215/TRIM22 modulates HCV replication by affecting NF-κB signaling. (A) The protein levels of p65 and phosphorylated p65 were determined by western blot in Con1b cells transfected with either miR-215 alone or combined with TRIM22. The activity of p65 was determined by calculating p-p65/p65 ratio. (B and C) The levels of HCV RNA and NS5A and NS3 proteins were determined in Con1b cells treated with NC or TNF-α. (D) The protein levels of p65 subunit of NF-κB (p65) and phosphorylated p65 (p-p65) were measured by western blot in Huh7.5.1 cells transfected with anti-miR-215 or cotransfected with anti-miR-215 and si-TRIM22. The activity of p65 was determined by calculating p-p65/p65 ratio. (E and F) The levels of HCV RNA and NS5A and NS3 proteins were analyzed in Huh7.5.1 cells treated with NC or PDTC. *p<0.05. HCV, hepatitis C virus; PDTC, pyrrolidinedithiocarbamate. |
Table 1
Predicted Target Genes of miR-215

References
1. Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013; 19:837–849.

2. Brjalin V, Salupere R, Tallo T, Kuznetsova T, Priimägi L, Tefanova V. Predictors of treatment response in patients with hepatitis C 1b genotype. Cent Euro J Med. 2013; 8:822–829.

3. Morozov VA, Lagaye S. Hepatitis C virus: morphogenesis, infection and therapy. World J Hepatol. 2018; 10:186–212.

4. Pol S, Parlati L. Treatment of hepatitis C: the use of the new pangenotypic direct-acting antivirals in “special populations”. Liver Int. 2018; 38:Suppl 1. 28–33.

5. Huang D, Koh C, Feurtado JA, Tsang EW, Cutler AJ. MicroRNAs and their putative targets in Brassica napus seed maturation. BMC Genomics. 2013; 14:140.

6. Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011; 3:83–92.
7. Piedade D, Azevedo-Pereira JM. MicroRNAs, HIV and HCV: a complex relation towards pathology. Rev Med Virol. 2016; 26:197–215.

8. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005; 309:1577–1581.

9. Liu X, Wang T, Wakita T, Yang W. Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology. 2010; 398:57–67.

10. Li S, Duan X, Li Y, Liu B, McGilvray I, Chen L. MicroRNA-130a inhibits HCV replication by restoring the innate immune response. J Viral Hepat. 2014; 21:121–128.

11. Lin Y, Jin Y, Xu T, Zhou S, Cui M. MicroRNA-215 targets NOB1 and inhibits growth and invasion of epithelial ovarian cancer. Am J Transl Res. 2017; 9:466–477.
12. Yao J, Zhang P, Li J, Xu W. MicroRNA-215 acts as a tumor suppressor in breast cancer by targeting AKT serine/threonine kinase 1. Oncol Lett. 2017; 14:1097–1104.

13. Vychytilova-Faltejskova P, Merhautova J, Machackova T, Gutierrez-Garcia I, Garcia-Solano J, Radova L, et al. MiR-215-5p is a tumor suppressor in colorectal cancer targeting EGFR ligand epiregulin and its transcriptional inducer HOXB9. Oncogenesis. 2017; 6:399.

14. Zang Y, Wang T, Pan J, Gao F. miR-215 promotes cell migration and invasion of gastric cancer cell lines by targeting FOXO1. Neoplasma. 2017; 64:579–587.

15. Wei Y, Sun J, Li X. MicroRNA-215 enhances invasion and migration by targeting retinoblastoma tumor suppressor gene 1 in highgrade glioma. Biotechnol Lett. 2017; 39:197–205.

16. Ishida H, Tatsumi T, Hosui A, Nawa T, Kodama T, Shimizu S, et al. Alterations in microRNA expression profile in HCV-infected hepatoma cells: involvement of miR-491 in regulation of HCV replication via the PI3 kinase/Akt pathway. Biochem Biophys Res Commun. 2011; 412:92–97.

17. Ozato K, Shin DM, Chang TH, Morse HC 3rd. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008; 8:849–860.

19. Yap MW, Stoye JP. TRIM proteins and the innate immune response to viruses. Adv Exp Med Biol. 2012; 770:93–104.

20. Barr SD, Smiley JR, Bushman FD. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 2008; 4:e1000007.

21. Gao B, Duan Z, Xu W, Xiong S. Tripartite motif-containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear-located RING domain. Hepatology. 2009; 50:424–433.

22. Yang C, Zhao X, Sun D, Yang L, Chong C, Pan Y, et al. Interferon alpha (IFNα)-induced TRIM22 interrupts HCV replication by ubiquitinating NS5A. Cell Mol Immunol. 2016; 13:94–102.

23. He Y, Weng L, Li R, Li L, Toyoda T, Zhong J. The N-terminal helix α(0) of hepatitis C virus NS3 protein dictates the subcellular localization and stability of NS3/NS4A complex. Virology. 2012; 422:214–223.

24. Yu S, Gao B, Duan Z, Xu W, Xiong S. Identification of tripartite motif-containing 22 (TRIM22) as a novel NF-κB activator. Biochem Biophys Res Commun. 2011; 410:247–251.

25. Waldron PR, Holodniy M. MicroRNA and hepatitis C virus—challenges in investigation and translation: a review of the literature. Diagn Microbiol Infect Dis. 2014; 80:1–12.

26. Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007; 449:919–922.

27. Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, et al. A cellular microRNA mediates antiviral defense in human cells. Science. 2005; 308:557–560.

28. Wang C, Chen Q, Li S, Li S, Zhao Z, Gao H, et al. Dual inhibition of PCDH9 expression by miR-215-5p up-regulation in gliomas. Oncotarget. 2017; 8:10287–10297.

29. Hou Y, Zhen J, Xu X, Zhen K, Zhu B, Pan R, et al. miR-215 functions as a tumor suppressor and directly targets ZEB2 in human non-small cell lung cancer. Oncol Lett. 2015; 10:1985–1992.

30. Mamdouh S, Khorshed F, Aboushousha T, Hamdy H, Diab A, Seleem M, et al. Evaluation of mir-224, mir-215 and mir-143 as serum biomarkers for HCV associated hepatocellular carcinoma. Asian Pac J Cancer Prev. 2017; 18:3167–3171.
31. Ali SA, Alahmady ZZ, Yamany HA, Abul-Fotouh AM. Serum expression levels of miR-141 and miR-215 for differentiation between liver cirrhosis, chronic hepatitis C and hepatocellular carcinoma patients. Microbiol Res J Int. 2017; 20:1–12.

32. Tissot C, Mechti N. Molecular cloning of a new interferon-induced factor that represses human immunodeficiency virus type 1 long terminal repeat expression. J Biol Chem. 1995; 270:14891–14898.

33. Singh R, Gaiha G, Werner L, McKim K, Mlisana K, Luban J, et al. Association of TRIM22 with the type 1 interferon response and viral control during primary HIV-1 infection. J Virol. 2011; 85:208–216.

34. Di Pietro A, Kajaste-Rudnitski A, Oteiza A, Nicora L, Towers GJ, Mechti N, et al. TRIM22 inhibits influenza A virus infection by targeting the viral nucleoprotein for degradation. J Virol. 2013; 87:4523–4533.

35. Ng TI, Mo H, Pilot-Matias T, He Y, Koev G, Krishnan P, et al. Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology. 2007; 45:1413–1421.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download