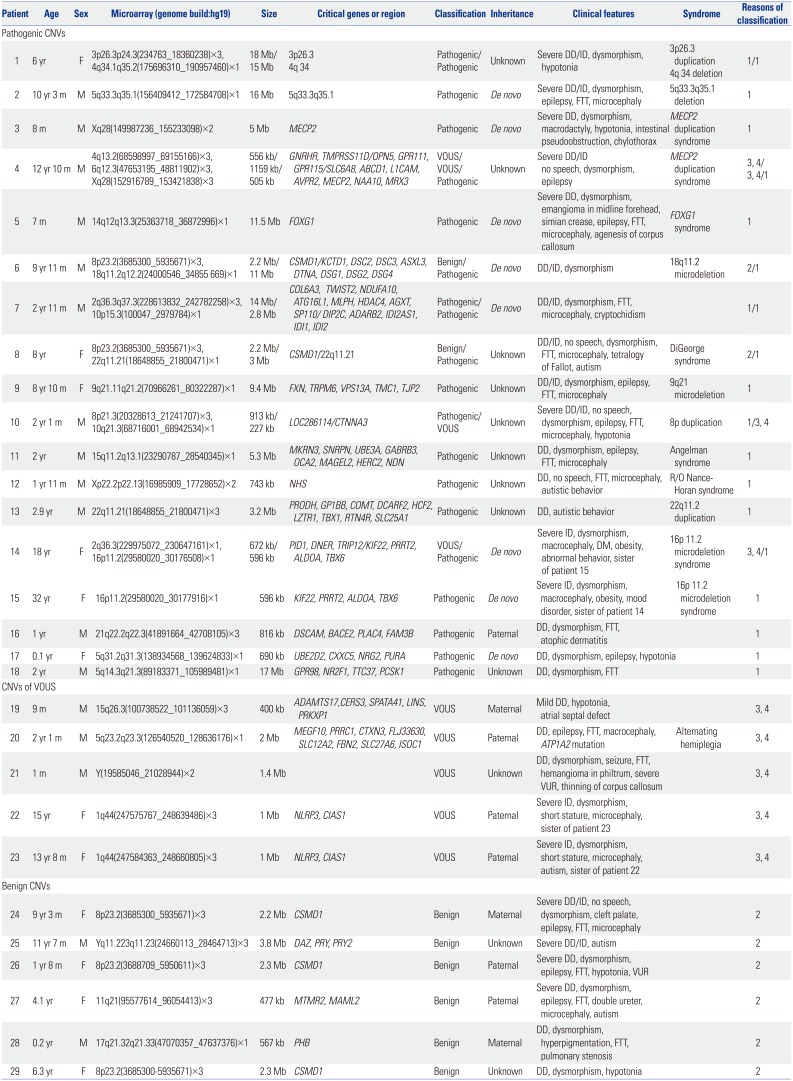
|
Pathogenic CNVs |
|
1 |
6 yr |
F |
3p26.3p24.3(234763_18360238)×3, 4q34.1q35.2(175696310_190957460)×1 |
18 Mb/15 Mb |
3p26.3 |
Pathogenic/Pathogenic |
Unknown |
Severe DD/ID, dysmorphism, hypotonia |
3p26.3 duplication |
1/1 |
|
4q 34 |
4q 34 deletion |
|
2 |
10 yr 3 m |
M |
5q33.3q35.1(156409412_172584708)×1 |
16 Mb |
5q33.3q35.1 |
Pathogenic |
De novo
|
Severe DD/ID, dysmorphism, epilepsy, FTT, microcephaly |
5q33.3q35.1 deletion |
1 |
|
3 |
8 m |
M |
Xq28(149987236_155233098)×2 |
5 Mb |
MECP2
|
Pathogenic |
De novo
|
Severe DD, dysmorphism, macrodactyly, hypotonia, intestinal pseudoobstruction, chylothorax |
MECP2 duplication syndrome |
1 |
|
4 |
12 yr 10 m |
M |
4q13.2(68598997_69155166)×3, 6q12.3(47653195_48811902)×3, Xq28(152916789_153421838)×3 |
556 kb/1159 kb/505 kb |
GNRHR, TMPRSS11D/OPN5, GPR111, GPR115/SLC6A8, ABCD1, L1CAM, AVPR2, MECP2, NAA10, MRX3
|
VOUS/VOUS/Pathogenic |
Unknown |
Severe DD/ID no speech, dysmorphism, epilepsy |
MECP2 duplication syndrome |
3, 4/3, 4/1 |
|
5 |
7 m |
M |
14q12q13.3(25363718_36872996)×1 |
11.5 Mb |
FOXG1
|
Pathogenic |
De novo
|
Severe DD, dysmorphism, emangioma in midline forehead, simian crease, epilepsy, FTT, microcephaly, agenesis of corpus callosum |
FOXG1 syndrome |
1 |
|
6 |
9 yr 11 m |
M |
8p23.2(3685300_5935671)×3, 18q11.2q12.2(24000546_34855 669)×1 |
2.2 Mb/11 Mb |
CSMD1/KCTD1, DSC2, DSC3, ASXL3, DTNA, DSG1, DSG2, DSG4
|
Benign/Pathogenic |
De novo
|
DD/ID, dysmorphism |
18q11.2 microdeletion |
2/1 |
|
7 |
2 yr 11 m |
M |
2q36.3q37.3(228613832_242782258)×3, 10p15.3(100047_2979784)×1 |
14 Mb/2.8 Mb |
COL6A3, TWIST2, NDUFA10, ATG16L1, MLPH, HDAC4, AGXT, SP110/DIP2C, ADARB2, IDI2AS1, IDI1, IDI2
|
Pathogenic/Pathogenic |
De novo
|
DD/ID, dysmorphism, FTT, microcephaly, cryptochidism |
|
1/1 |
|
8 |
8 yr |
F |
8p23.2(3685300_5935671)×3, 22q11.21(18648855_21800471)×1 |
2.2 Mb/3 Mb |
CSMD1/22q11.21 |
Benign/Pathogenic |
Unknown |
DD/ID, no speech, dysmorphism, FTT, microcephaly, tetralogy of Fallot, autism |
DiGeorge syndrome |
2/1 |
|
9 |
8 yr 10 m |
F |
9q21.11q21.2(70966261_80322287)×1 |
9.4 Mb |
FXN, TRPM6, VPS13A, TMC1, TJP2
|
Pathogenic |
Unknown |
DD/ID, dysmorphism, epilepsy, FTT, microcephaly |
9q21 microdeletion |
1 |
|
10 |
2 yr 1 m |
M |
8p21.3(20328613_21241707)×3, 10q21.3(68716001_68942534)×1 |
913 kb/227 kb |
LOC286114/CTNNA3
|
Pathogenic/VOUS |
Unknown |
Severe DD/ID, no speech, dysmorphism, epilepsy, FTT, microcephaly, hypotonia |
8p duplication |
1/3, 4 |
|
11 |
2 yr |
M |
15q11.2q13.1(23290787_28540345)×1 |
5.3 Mb |
MKRN3, SNRPN, UBE3A, GABRB3, OCA2, MAGEL2, HERC2, NDN
|
Pathogenic |
Unknown |
DD, dysmorphism, epilepsy, FTT, microcephaly |
Angelman syndrome |
1 |
|
12 |
1 yr 11 m |
M |
Xp22.2p22.13(16985909_17728652)×2 |
743 kb |
NHS
|
Pathogenic |
Unknown |
DD, no speech, FTT, microcephaly, autistic behavior |
R/O Nance-Horan syndrome |
1 |
|
13 |
2.9 yr |
M |
22q11.21(18648855_21800471)×3 |
3.2 Mb |
PRODH, GP1BB, COMT, DCARF2, HCF2, LZTR1, TBX1, RTN4R, SLC25A1
|
Pathogenic |
Unknown |
DD, autistic behavior |
22q11.2 duplication |
1 |
|
14 |
18 yr |
F |
2q36.3(229975072_230647161)×1, 16p11.2(29580020_30176508)×1 |
672 kb/596 kb |
PID1, DNER, TRIP12/KIF22, PRRT2, ALDOA, TBX6
|
VOUS/Pathogenic |
De novo
|
Severe ID, dysmorphism, macrocephaly, DM, obesity, abnormal behavior, sister of patient 15 |
16p 11.2 microdeletion syndrome |
3, 4/1 |
|
15 |
32 yr |
F |
16p11.2(29580020_30177916)×1 |
596 kb |
KIF22, PRRT2, ALDOA, TBX6
|
Pathogenic |
De novo
|
Severe ID, dysmorphism, macrocephaly, obesity, mood disorder, sister of patient 14 |
16p 11.2 microdeletion syndrome |
1 |
|
16 |
1 yr |
M |
21q22.2q22.3(41891664_42708105)×3 |
816 kb |
DSCAM, BACE2, PLAC4, FAM3B
|
Pathogenic |
Paternal |
DD, dysmorphism, FTT, atophic dermatitis |
|
1 |
|
17 |
0.1 yr |
F |
5q31.2q31.3(138934568_139624833)×1 |
690 kb |
UBE2D2, CXXC5, NRG2, PURA
|
Pathogenic |
De novo
|
DD, dysmorphism, epilepsy, hypotonia |
|
1 |
|
18 |
2 yr |
M |
5q14.3q21.3(89183371_105989481)×1 |
17 Mb |
GPR98, NR2F1, TTC37, PCSK1
|
Pathogenic |
Unknown |
DD, dysmorphism, FTT |
|
1 |
|
CNVs of VOUS |
|
19 |
9 m |
M |
15q26.3(100738522_101136059)×3 |
400 kb |
ADAMTS17, CERS3, SPATA41, LINS, PRKXP1
|
VOUS |
Maternal |
Mild DD, hypotonia, atrial septal defect |
|
3, 4 |
|
20 |
2 yr 1 m |
M |
5q23.2q23.3(126540520_128636176)×1 |
2 Mb |
MEGF10, PRRC1, CTXN3, FLJ33630, SLC12A2, FBN2, SLC27A6, ISOC1
|
VOUS |
Paternal |
DD, epilepsy, FTT, macrocephaly, ATP1A2 mutation |
Alternating hemiplegia |
3, 4 |
|
21 |
1 m |
M |
Y(19585046_21028944)×2 |
1.4 Mb |
|
VOUS |
Unknown |
DD, dysmorphism, seizure, FTT, hemangioma in philtrum, severe VUR, thinning of corpus callosum |
|
3, 4 |
|
22 |
15 yr |
F |
1q44(247575767_248639486)×3 |
1 Mb |
NLRP3, CIAS1
|
VOUS |
Paternal |
Severe ID, dysmorphism, short stature, microcephaly, sister of patient 23 |
|
3, 4 |
|
23 |
13 yr 8 m |
F |
1q44(247584363_248660805)×3 |
1 Mb |
NLRP3, CIAS1
|
VOUS |
Paternal |
Severe ID, dysmorphism, short stature, microcephaly, autism, sister of patient 22 |
|
3, 4 |
|
Benign CNVs |
|
24 |
9 yr 3 m |
F |
8p23.2(3685300_5935671)×3 |
2.2 Mb |
CSMD1
|
Benign |
Maternal |
Severe DD/ID, no speech, dysmorphism, cleft palate, epilepsy, FTT, microcephaly |
|
2 |
|
25 |
11 yr 7 m |
M |
Yq11.223q11.23(24660113_28464713)×3 |
3.8 Mb |
DAZ, PRY, PRY2
|
Benign |
Unknown |
Severe DD/ID, autism |
|
2 |
|
26 |
1 yr 8 m |
F |
8p23.2(3688709_5950611)×3 |
2.3 Mb |
CSMD1
|
Benign |
Paternal |
Severe DD, dysmorphism, epilepsy, FTT, hypotonia, VUR |
|
2 |
|
27 |
4.1 yr |
F |
11q21(95577614_96054413)×3 |
477 kb |
MTMR2, MAML2
|
Benign |
Paternal |
Severe DD, dysmorphism, epilepsy, FTT, double ureter, microcephaly, autism |
|
2 |
|
28 |
0.2 yr |
M |
17q21.32q21.33(47070357_47637376)×1 |
567 kb |
PHB
|
Benign |
Maternal |
DD, dysmorphism, hyperpigmentation, FTT, pulmonary stenosis |
|
2 |
|
29 |
6.3 yr |
F |
8p23.2(3685300-5935671)×3 |
2.3 Mb |
CSMD1
|
Benign |
Unknown |
DD, dysmorphism, hypotonia |
|
2 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download