Abstract
The intestinal microbiota is a complex ecosystem consisting of various microorganisms that expands human genetic repertoire and therefore affects human health and disease. The metabolic processes and signal transduction pathways of the host and intestinal microorganisms are intimately linked, and abnormal progression of each process leads to changes in the intestinal environment. Alterations in microbial communities lead to changes in functional structures based on the metabolites produced in the gut, and these environmental changes result in various bacterial infections and chronic enteric inflammatory diseases. Here, we illustrate how antibiotics are associated with an increased risk of antibiotic-associated diseases by driving intestinal environment changes that favor the proliferation and virulence of pathogens. Understanding the pathogenesis caused by antibiotics would be a crucial key to the treatment of antibiotic-associated diseases by mitigating changes in the intestinal environment and restoring it to its original state.
The intestinal microbiota plays beneficial roles in many physiological processes of the host. It extracts energy and nutrition from food, protects against enteropathogens, and supports development and maintenance of the host immune system.123 The biodiversity of the intestinal microbiota among individuals implies that it sustains a homeostatic equilibrium state against a decrease in its composition and function.45 The particular interrelationship between the intestinal microbiota and the host is a product of long-term coexistence and evolution.567 Dysbiosis, a disruption of microbial composition by various stresses, has been implicated in inflammatory bowel disease (IBD), colon cancer, obesity, asthma, and other diseases.189 The first step in treatment of these diseases is to understand the symbiotic relationship between the intestinal microbiota and its host. Here, we review and outline studies that have discovered how antibiotics change microbial composition, resultant physical and chemical changes in the body, and how such changes become a trigger for disease. Finally, we discuss recent progress toward approaches aimed at restoring a disturbed ecosystem.
Vertebrates host a dense microbial community of bacteria, viruses, and fungi, namely the microbiota, in organs containing mucous membranes, such as the oral cavity and intestines. In healthy individuals, Proteobacteria, Bacteroidetes, and Archaebacteria are considered the major bacterial taxa.10 Residing in the intestines, these diverse microorganisms develop elaborate ecological networks through interactions with other bacteria to obtain nutrients required for their colonization and proliferation.34111213 Host-microbial and microbial-microbial interactions establish an equibrilium state of microbial composition in the intestinal tract.61014 The complete intestinal microbiota maintains intestinal symbiosis by suppressing invasion of microorganisms from the outside and regulating the excessive proliferation of microorganisms that are present in the intestines in small numbers. When the intestinal microbial communities collapse or become unbalanced due to a variety of causes, such as antibiotics, chemical toxic substances, pathogen infections, and drastic changes in dietary habits, the immune responses of the host act abnormally leading to IBDs.6815 In particular, excessive dosing of antibiotics elicits the loss of naturally occurring intestinal microbiota. Such loss increases the numbers of yeasts, such as Candida albicans, and bacteria, such as Proteus, Staphylococcus, and Clostridium difficile (C. difficile), that normally exist at low numbers, leading to depression of digestive functions or the occurrence of intestine-related diseases.891617 It is understood that external stimuli initially induce disturbances in the intestinal environment that result in one of four states−resistance, resilience, redundancy, or dysbiosis−depending on whether the disturbance is overcome by the intestinal microbial ecosystem.41214181920 When the intestinal microbiota responds appropriately to any fluctuations and is recovered to its original state before being stressed by environmental perturbations, the intestinal environmental state is considered to be resistance.7 However, in cases where the stress is very powerful, alterations in the community at the level of genera or species and loss of functions occur, and the intestinal environment begins to change in the following three directions: If the effect is insignificant, the intestinal ecosystem enters a resilience state with a microbial community similar to the original one through reshaping toward the initial state. In the state of redundancy, growth of bacteria different from those in the initial state increases such that the diversity of the bacteria increases, although the genes do not undergo functional changes. In this case, proteins and metabolites similar to those in the initial state, in terms of function, are produced.121821 Resistance, resilience, and redundancy are attributes that appear when the intestinal microbiota is strong and shows functional recovery.141518 In contrast, dysbiosis refers to a state where irreversible changes occur in the intestinal microbiota with variations and functional damage at the level of genes or proteins, eventually leading to disturbances of responses and the immune system in intestinal epithelial cells, together with changes in intestinal metabolites.
Antibiotics can be a very powerful factor causing imbalance of the intestinal microbiota.222324 In 1954, Bohnhoff, et al. noticed that mice that were given streptomycin were easily infected by Salmonella enterica serovar Enteritidis and introduced the concept that intestinal microbiota could suppress the growth of bacteria that invade mice from the outside through colonization resistance.25 Direct interaction of intestinal microbiota with bacteria and competition for intestinal nutrients are direct methods of inhibiting the intestinal colonization of pathogens. However, dosing with antibiotics reduces the diversity and abundance of intestinal microbiota, leading to a reduction in the competitive exclusion ability.262728 Indirectly, this destroys the community structure, thereby disturbing the interactions among microbial species and the complementary systems of nutrient metabolic pathways, resulting in widespread fluctuations in the intestinal environment. These changes are not fully reversed, even after several months of discontinuation of dosing.1622 Eventually, the antibiotic-induced dysbiosis of the intestinal microbiota affects the development and regulation of the immune system and increases the risk of intestine-related diseases, such as IBDs and infectious diseases, in addition to diverse immunity-related disorders, such as allergic or atopic skin diseases and type 1 diabetes.162829 In an experiment where mice were treated with antibiotics at sub-therapeutic concentrations, changes in the composition of intestinal microbiota were associated with changes in total body weight, body fat content, bone density, production of short-chain fatty acids (SCFAs), and hepatic fatty acid metabolism. To understand the effects of antibiotics on the homeostasis of the immune system, we first need to understand how antibiotics extensively change the intestinal microbial ecosystem. Antibiotics are generally administered to kill specific microorganisms; however, since most antibiotics have a wide range of effects, they also affect related microorganisms. These effects are imprinted in the intestinal environment for several months after discontinuation of the dosing.19233031 The effects of the antibiotics on the taxonomic composition of intestinal microbiota vary among individuals, and symptoms, such as reduction of the diversity of bacteria, a decrease in homology, and relative ex-cessive increases of certain species, are restored or persist. Other symptoms may also occur, depending on differences in characteristics among individuals.
Studies on the effect of exposure to antibiotics immediately after birth show that the abundance and diversity of intestinal microbiota change regardless of the kind of antibiotic.3233 The intestinal microbiota of preterm infants born before 33 weeks is about 10 times smaller than that of infants born at term. In addition, exposure to diverse antibiotics immediately after birth also leads to marked differences in the initial formation of microbial communities. In particular, the abundance and diversity of the intestinal microbiota decreases rapidly with exposure to meropenem, cefotaxime, and ticarcillin-clavulanate. Unlike other antibiotics, administration of vancomycin and gentamicin does not change the diversity very much, but increases in the expression of genes related to resistance to these antibiotics are observed. Upregulation of such genes shows a close relationship with increases in certain microorganisms. In particular, ticarcillin-clavulanate and ampicillin are associated with a marked increase in Klebsiella pneumoniae, together with an increase in gene expression related to resistance to the antibiotics.34
Anaerobic bacteria account for a large proportion of the intestinal microbiota and play an important role in the development of intestinal immunity.1526323335 These organisms produce volatile fatty acids, such as butyrate, which activate im-mune cells to help maintain a healthy gut. Clindamycin, a br-oad-spectrum antibiotic against anaerobic bacteria, is released from bile after administration and accumulates at high concentrations in the feces, affecting the composition of the intestinal microbiota for a long time, even after discontinuation of dosing.22 Clindamycin is known to have the greatest effect on the function of intestinal microbiota that shows resistance to the pathogenic bacteria C. difficile. Studies of the effects of clarithromycin, metronidazole, and omeprazole on the composition of the pharyngeal and fecal bacterial taxa show that these antibiotics may affect 30% or more of the microbiota composition, and although the microbiota may partially recover, the effects can persist for at least 4 years after exposure.33 In addition, analysis of the changes in microbiota between before and after treatment of patients with fluoroquinolone and β-lactam antibiotics for 1 week showed that the 16S rRNA copy number did not change or increased, while changes in the composition of microbiota and a reduction in the diversity were observed. In particular, an increase in the ratio of Bacteroidetes to Firmicutetes was observed, and the distribution of the bacterial taxa tended to be simplified.26
Even ciprofloxacin, which exhibits relatively low activity ag-ainst anaerobic bacteria, can have a profound effect on the composition of intestinal microbiota.3637 Ciprofloxacin was prescribed for 5 days to patients who had not taken the antibiotic previously and changes in the intestinal microbiota between 60 days before dosing and after dosing and between after dosing and 6 months after discontinuation of dosing were analyzed. The results showed that the diversity of bacterial taxa decreased by approximately one-third, and the taxonomic ab-undance, diversity, and uniformity of the intestinal microbiota decreased.3638 The results of another study indicated that the diversity of the bacterial taxa had already begun to decrease in the early stages of exposure to the antibiotic, and when differences in responses among individuals and the degree and time of recovery of the microbiota were measured, universal decreases in Ruminococcaceae were observed, in addition to the rapid destruction of bacterial diversity. Although some compositional changes persisted, even after 6 months had pass-ed, most of the compositions showed a tendency to recover almost completely by 1 month after dosing. These differences were shown to be attributable to the diversity of intestinal microbiota in the early stage, indicating that the diversity of the bacterial taxa of intestinal microbiota plays an important role in the subsequent recovery of diversity to the redundancy state.537 In addition, re-exposure to the same amount of ciprofloxacin 6 months later showed similar effects on the structure of intestinal microbiota, although there was a tendency for less efficient recovery.36
Antibiotics trigger the proliferation of intestinal pathogenic bacteria22 due to the ability of infectious bacteria to effectively exploit the disorder that arises when the intestinal microbiota has collapsed.1039 Although there are still many questions about the causes of increased bacterial infections in the intestines after antibiotic treatment, the most interesting facts revealed thus far are described below (Fig. 1).
In the intestinal environment, obligatory anaerobic bacteria such as Bacteroidia and Clostridia lack genes related to aerobic respiration and grow through the fermentation of amino acids or polysaccharides (Fig. 1A). Imbalance of the intestinal microbiota induces intestinal inflammatory responses, and the most important environmental change in this regard is the increase in reactive nitrogen species (RNS) and reactive oxygen species (ROS). In the case of patients suffering from IBD, expression of inducible nitric oxide synthase in the intestinal epithelium increases and nitric oxide (NO−) concentrations in the lumen of the colon increases. NO− reacts with superoxide radicals (O2−) to give peroxynitrite (ONOO−), thereby producing nitrate (NO3−), or oxidizes organic sulfides and tertiary amines into S-oxides and N-oxides, respectively. Unlike obligatory anaerobic bacteria, Enterobacteriaceae are capable of anaerobic respiration using nitrate, S-oxides, and N-oxides as the final electron transport receptors.40 ROS and RNS produced by the host's immune system can be utilized by facultative anaerobic bacteria such as Escherichia coli (E. coli) and aerobic bacteria.4142 E. coli is present in small numbers in healthy intestines, but its levels tend to increase significantly in streptomycin-treated mice (Fig. 1B). An E. coli mutant lacking moaA, a gene necessary for the biosynthesis of the molybdenum cofactor that is absolutely necessary for the activity of nitrate reductases, S-oxide reductases, and N-oxide reductases, tends to show a reduction in intestinal proliferation in mice with inflammation induced by dextran sulfate sodium.42 Salmonella also undergo aerobic respiration using the ROS produced by neutrophils and tetrathionate (S4O62-), an electron transfer compound produced in the oxidation pathway of hydrogen sulphide (H2S) produced by microorganisms (Fig. 1B).42 In a study of the pathogenesis of Vibrio cholerae under the oxidative stress induced by various antibiotics, overgrowth of Enterobacteriaceae and Enterococci was observed after administration of streptomycin.43 This study showed that changes in the community of intestinal microbiota caused by the antibiotic lead to increases in ROS in the intestinal environment. Atypical E. coli harboring extra catalase (katE) are adapted to this environment and excessively proliferate resulting in a temporary decrease in the ROS concentration, and cholera bacteria can effectively use such conditions (Fig. 1E).
Microorganisms produce and consume products through different metabolic pathways, and the ecosystem therefore consists of highly sophisticated networks.4445 The Archaebacteriae Methanobrevibacter smithii and Bacteroides thethaiotaomicron can be more effectively established in sterile rats together than singly due to cooperation in polysaccharide metabolism through the pathway that converts fructan to acetate and the resultant formate.46 In addition, results indicating that Bifidobacterium adolescentis, which degrades macromolecular carbohydrates, is capable of providing the substrates lactate and acetate to bacteria that produce butyrate in the intestines demonstrate the ability of bacteria to use each other to overcome conditions of insufficient metabolic pathways.47
Antibiotics have been reported to alter the intestinal microbiota involved in carbohydrate metabolism, thereby increasing the intestinal concentrations of carbohydrates essential for the proliferation of infectious bacteria.48 Most intestinal resident bacteria and pathogenic bacteria can utilize intestinal sialic acid (Neu5Ac) as a nutrient (Fig. 1C). Bacteroides thetatiotaomicron (B. thetatiotaomicron), one of the representative commensal bacteria, possesses a sialidase enzyme capable of degrading glycoconjugates present in the mucosa in order to produce sialic acid but the metabolic pathway that consumes it is incomplete.49 In contrast, Salmonella enterica serovar Typhimurium (S. Typhimurium hereafter) and C. difficile have a nan operon, which is necessary to use sialic acid, but do not have sialidase, which is required to produce sialic acid from the intestinal mucosa. When B. thetatiotaomicron was transferred to germ-free mice and the degree of sialic acid metabolism by S. Typhimurium and C. difficile in these mice was compared with that in untreated mice, increases in the expression of nanE (a gene in the sialic acid degradation pathway) and the fuc1 operon (fucose metabolizing gene cluster) were observed in S. Typhimurium proliferating in B. thetatiotaomicron mice. In the case of C. difficile, increases in the expression of nanA and nanE genes were observed. In addition, when normal mice were treated with streptomycin relatively more sialic acid was produced and increases in the expression of genes related to sialic acid metabolism were identified in the group treated with the antibiotic.49 Another study reported that C. difficile is capable of proliferating in the intestine using the succinate-butyrate metabolic pathway (Fig. 1D). In the presence of B. thetatiotaomicron, the pathway by which C. difficile metabolizes succinate, a product of fermentation by B. thetatiotaomicron, into butyrate is further induced.50 In environments with excess polysaccharide, B. thetatiotaomicron produces a high concentration of succinate and C. difficile produces butyrate. In addition, the amount of succinate present in the intestines of mice with normal bacterial flora increased following antibiotic treatment, and mutant strains deficient in the ability to use succinate due to loss of the succinate transporter showed decreased intestinal proliferation.50 In fact, B. thetatiotaomicron is a normal intestinal bacterial flora and a beneficial bacterium with the ability to metabolize diverse carbohydrates. Therefore, the production of succinate also occurs under the condition of normal bacterial flora.51 However, in this study, levels of the SCFAs acetate and butyrate decreased, while that of succinate increased, in intestines where diarrhea was induced by treatment with antibiotics or polyethylene glycol, indicating that the increase in succinate due to changes in intestinal gluconeogenesis caused by antibiotics can be one of the various causes that promote the proliferation of C. difficile. The use of succinate is also observed in Citrobacter rodentium (C. rodentium) (Fig. 1D). C. rodentium is known to initiate expression of the pathogenic factor genes ler, espA, eae, nleAr, and stx2, which are the locus of the enterocyte effacement genes essential for intestinal infections, by recognizing succinate and regulating the transcription factor Cra.52
Intestinal inflammatory responses help S. Typhimurium spread to the lumen of the large intestine, and migration to the colon facilitates feces-oral cavity transfer to other highly susceptible hosts.255354 S. typhimurium, which induces salmonellosis in humans, has been reported to be amplified in mice treated with vancomycin and streptomycin.16 In particular, excessive use of antibiotics during post-operative convalescence may cause recurrence of bacterial infections and pathological symptoms. Known pathogenic factors for non-typhoid Salmonella, such as S. typhimurium, include the invasion-associated type III secretion system (T3SS-1) necessary for the pathogen to enter the intestinal epithelial cells, and the second type III secretion system (T3SS-2) necessary for survival of the pathogen in the tissues. The increase in proliferation of S. Typhimurium in the intestines is attributed to increases in respiratory electron transport components generated during the host's inflammatory responses (Fig. 1B). For instance, S. Typhimurium can form nitrate, a substrate for anaerobic respiration, using RNS, one of the outcomes of inflammation.55 Other examples are explained by the use of SCFAs (Fig. 2A). SCFAs such as butyrate, acetate, and propionate are produced by the anaerobic bacteria present in the large intestine and are used for barrier functions such as IL-8 secretion and mucus production by intestinal epithelial cells, the tight junction between intestinal cells, and the activation of intestinal cells.445657 They are also used as an energy source for intestinal cells. In addition, butyrate plays a role in regulating hypoxia-inducible factor (HIF), a transcription factor that regulates the barrier function, mucus production, and defends against pathogens of intestinal epithelial cells.4557 The epithelium of the large intestine is relatively hypoxic because it is located between the intestinal lumen with low oxygen partial pressure and the lamina propria with high oxygen concentration. SCFAs promote O2 consumption of human intestinal cells to maintain the intestinal environment anaerobically and stabilize HIF-1α, a subunit of HIF. After treatment with antibiotics, staining with the O2- sensitive dye pimonidazole was lost, and the activity of HIF-1α was decreased. In addition, oral administration of butyrate resulted in an increase in HIF-1α concentration and buytrate concentration-dependent recovery of the barrier effect of intestinal cells.45
Antibiotic treatment increases oxygen production in the colon epithelial cells, increasing the oxygen content in the intestinal lumen. A decrease in Clostridia, a producer of butyrate, is expected to increase the oxidation reactions of the intestinal cells and promote the diffusion of oxygen into the intestinal lumen (Fig. 2). Oxygen is the only respiratory electron transport receptor with greater oxidation-reduction potential than nitrate, and is more effectively used by aerobic or facultative anaerobic microorganisms. The oxygen partial pressure, which is about 100 mm Hg in the basal layer, becomes 20−40 mm Hg in the mucus layer and reaches almost 0 mm Hg in the lumen, leading to hypoxia.58 Therefore, changes in the intestinal oxygen partial pressure caused by antibiotic treatment are the first obstacle to the survival of intestinal microbiota and can be a very important factor in the induction of infectious disease. Colonic cells oxidize butyrate to form CO2, leading to a hypoxic (<7.6 mm Hg or <1% O2) state. However, in the case of neonatal mice, since butyrate, which is a metabolite of intestinal microbiota, is not present, energy is obtained by producing lactate from glucose. This process increases the oxygenation of intestinal cells.45 For respiration using oxygen, Salmonella uses cytochrome bd oxidase produced by cydA and cytochrome bd-II oxidase produced by cyxB. Salmonella can proliferate using the cytochrome bd-II oxidase at a low oxygen partial pressure (0.8%) (Fig. 2B).54 Moreover, treatment with tributyrin rescued the large intestine of streptomycin-treated mice from the hypoxia state and increased the butyrate concentration of the cecum.
The most serious gut-associated disease caused by antibiotics is pseudomembranous, which is a typical antibiotic-associated diarrhea caused by an increase in C. difficile. C. difficile infection (CDI) is one of the most common pathogenic infections, and is particularly prevalent among patients continuously taking antibiotics.485960 CDI was first recognized in advanced countries such as the United States and European countries where many cases are observed. The treatment and recurrence of this disease occur repeatedly, and since recurrent cases cannot be easily treated with general antibiotics, this bacterium is classified as a serious disease causative organism.59 Control and complete treatment of C. difficile is hard because its spores can survive several years, even in alcohol.48 Although there is a continuously increasing trend in the number of CDI patients in foreign countries the number of cases in South Korea is still small, although collective outbreaks are considered possible.
Approaches such as developing antibiotics with a new active mechanism, targeting pathogenic factors, or using native microorganisms such as probiotics, have been attempted.6162 However, such developments or attempts still generate many controversies in terms of safety or effectiveness. In the following section, representative non-antibiotic treatment methods used to treat C. difficile and other gut-associated diseases are introduced.
Fecal microbiota transplantation (FMT), a method that involves injecting the fecal microorganisms of healthy persons who are highly likely to have similar structures of intestinal microbiota to those of the patient in order to restore normal bacterial flora in the intestines, has been shown to be effective in diverse clinical trials. Transplantation of the fecal microbiota leads to re-establishment of the patients' intestinal environment with the composition of the intestinal microbiota of the donor, eventually inducing relative control of C. difficile and thereby enabling effective treatment.6364 This method is very effective and has been reported to show cure rates in the range of 80−100%, according to the number of times of FMT administration. It is very efficient in that the provision of healthy intestinal microbiota enables acquisition of the ability to resist the secondary problem of infection with other pathogens such as vancomycin-resistant Enterococcus and carbapenem-resistant Enterobacteriaceae.486465 FMT appears to be an important alternative to antibiotics that destroy important commensal bacteria and provides colonization resistance, such as niche exclusion for pathogens, the production of antimicrobials, and activation of the immune system of the mucous layer.64
Recent studies have attempted to control Salmonella infections using bacterial flora, the taxa and characteristics of which were identified among bacteria isolated from the intestines.62 The Microbial Ecosystem Therapeutic (MET-1) is an ecological system of 33 kinds of microorganisms isolated from human feces, consisting of Actinobacteria (mainly Bifidobacterium spp.), Bacteroidetes, Firmicutes, and Proteobacteria. In MET-1-treated mice, body weight loss due to S. Typhimurium infection was observed, as well as a decrease in serum cytokines, NF-κB nuclear staining, and neutrophil infiltration in the cecum. In addition, ZO-1, a tight junction protein, was preserved in the cecum, cellular localization of claudin-1 decreased, and S. Typhimurium translocation to the spleen decreased. However, there was no change in the colony forming units of Salmonella in the intestine. Therefore, MET-1 was th-ought to regulate systemic infections through maintenance of the tight junction to control access to the systemic circulation.62
Despite many attempts and positive results, questions still remain about the safety of FMT. The interactions between pa-thogenic bacteria and microorganisms are not simple and related information is still insufficient. In addition, industrial use of FMT is increasing, and the use of FMT by patients who have not been definitely diagnosed with CDI may cause other problems, and there is the possibility of the existence of other pathogens in the donor's intestinal microbiota. Therefore, the use of FMT requires strict criteria regarding the optimized composition of intestinal microbiota, based on the composition and functions of microorganisms that fit the causative disease to be treated, the age of the recipient, and the stage of disease progression.63 Obtaining a verified bacterial flora through total examination of the health condition of the donor, determination of whether any genetic disease is latent, and detailed analysis of the composition of intestinal microbiota is very important.64
The focus on probiotics began in 1908 when Metchnikoff reported the relationship between fermented food and longevity, explaining that intestinal microbiota develop the mucosal immune system and can prevent the invasion of infectious bacteria. Thereafter, diverse bacterial taxa were tested under various experimental conditions to determine whether probiotics could treat a variety of gut-associated diseases such as Crohn's disease and ulcerative colitis, Irritable Bowel Syndrome, CDI, infectious diarrhea, and necrotizing enterocolitis.35616667 The idea that probiotics could improve or prevent diarrhea began with the notion that these gut-associated diseases were caused by the collapse of “colonization resistance” due to the absence of normal bacterial flora. In many studies, probiotics have been used to treat gut-associated diseases through activation of the immune system, competition for settlement sites in the intestinal cells, and the production of bacteriocin. These effects vary with the form and causes of diarrhea, such as viral diarrhea, antibiotic-related diarrhea, or traveler's diarrhea.
Probiotics are known to be very effective for the treatment of antibiotic-associated diarrhea, with Saccharomyces boulardii (S. boulardii), E. coli Nissle 1917, Lactobacillus, and Bifidobacterium as the main focus of research.6668 Positive effects were identified under conditions where lactic acid bacteria such as Lactobacillus GG (LGG), S. boulardii, E. faecium, Lactobacillus acidophilus (L. acidophilus), and Lactobacillus bulgaris were taken together with diverse types of antibiotics.35 When healthy adults were instructed to take erythromycin and LGG, a reduction in the duration of diarrhea symptoms from 8 days to 2 days was observed, and the incidence of related symptoms such as abdominal pain was reduced from 39% to 23%. In a pediatric study conducted with approximately 200 children who were administered antibiotics, prescription of LGG reduced the incidence of diarrhea from 26% to 8% and reduced the period of diarrhea from 5.88 days to 4.7 days.67 In addition, L. plantarum 299v administered together with oral antibiotics alleviated the symptoms of diseases that occurred when the use of antibiotics was discontinued, and L. rhamnosus prevented diarrhea caused by antibiotic treatment.68 S. boulardii is able to degrade C. difficile toxin A and toxin B by releasing a 54 kDa protease. In a study conducted with 138 hospital patients in which patients who were randomly prescribed with the probiotic strain were compared with placebo patients, only 2.9% of those who took the probiotics were found to be C. difficile toxin-positive, compared with 7.25% of placebo patients.61 When all fecal samples were examined, only 46% of the group that took probiotics showed C. difficile toxin-positive reactions compared with 78% of the placebo group. In addition, the efficacy of probiotics as adjuvant therapeutic agents has also been demonstrated. In a study conducted with 124 adult Clostridium difficile-associated disease patients, approximately 60% of patients prescribed with S. boulardii together with antibiotics showed relief of infection symptoms. In addition, S. boulardii has also shown relative effects for the prevention of secondary diseases in patients at risk of recurrence of CDI,6167 although this is controversial.
The results of studies conducted thus far indicate that the mechanisms through which probiotics can treat diseases are quite diverse, and information on which mechanisms individual probiotics induce has not been fully verified. These mechanisms vary with the types of bacterial taxa used as probiotics and the kind of experimental disease, and the known effects mainly include maintenance of interactions between the host and microorganisms, removal of bacteria, mucus secretion from goblet cells, control of the epithelial barrier function of the intestinal cells, production of anti-bacterial factors such as lactic acid, hydrogen peroxide, and bacteriocin, and activation of the acquired immune system of the host.20356768 However, the different mechanisms used to produce beneficial effects make the selection of probiotics difficult. Essentially, to verify the effects it is necessary to identify whether the probiotics would survive, as well as the surface proteins and bacteriocin produced. In addition, in order to use probiotics for diverse diseases, both the disease and the mechanisms of the probiotics should be accurately delineated. Careful selection of fully understood and proven probiotics may provide an alternative therapy to improve health and replace antibiotics for primary therapeutic purposes.
Antibiotic-associated diarrhea occurring in patients who take antibiotics emphasizes the importance of a balanced intestinal microbiota. The effects of antibiotics on intestinal microbiota are in fact clues to understanding which molecular substances or mechanisms each infectious bacterium can ef-fectively utilize in the changed environment in order to grow and cause disease in the host environment. Recent studies have provided information on how antibiotics can alter the intestinal environment, how harmful bacteria and beneficial bacteria react, and how pathogenic bacteria use these environments. Pathogens exploit the sugars, radicals, and oxygen occurring as a result of disruption of intestinal microbiota and the host inflammatory response. Application of FMT and probiotics for eradication of gastrointestinal diseases and enteropathogens exhibits the potential to restore the degraded ecosystem and protection against colonization and proliferation of enteropathogens. This new research has given us a greater understanding and new directions when considering future approaches to treat antibiotic-related infectious diseases.
Figures and Tables
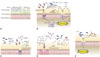 | Fig. 1Pathogens exploit the antibiotic-induced inflammatory conditions. Pathogens use sugars and inorganic compounds generated by the intestinal microbiota as carbon or energy sources and perform anaerobic respiration in the inflammatory conditions caused by antibiotics. (A) When the distribution of intestinal microorganisms is in a stable state, the invasion of pathogenic bacteria is suppressed by antimicrobial substances produced from intestinal bacteria and host cells and the inflammation is suitably controlled. (B) In an inflammatory condition, the colonization of E. coli and Salmonella expands through anaerobic respiration using ROS and RNS, which are released by DUOX2 and iNOS in epithelial cells. Hydrogen sulfide derived from sulfate-reducing bacteria such as Desulfovibrio spp. is converted to thiosulfate during cellular respiration in colonic epithelial cells. ROS generated by neutrophils convert the thiosulfate into tetrathionate that can be used as an electron acceptor. During this process, the generated tetrathionate boosts the growth of S. Typhimurium through tetrathionate respiration that converts tetrathionate to thiosulfate. E. coli reduces nitrate to nitrite through nitrate respiration. (C) Bacteroides thetaiotaomicron decomposes mucosal glycoconjugates to produce sialic acid. EHEC and Salmonella can use the sialic acid as a carbon source. Inflammatory conditions lead to release of fucose from host glycan and the liberated fucose is subsequently consumed by pathogens. As an example, EHEC are known to regulate the expression of virulence genes by sensing the fucose. (D) C. difficile, C. rodentium, and EHEC utilize succinate, which is produced by other intestinal microorganisms. SCFAs excreted during polysaccharide metabolism by aerobic bacteria and butyrate, propionate, and acetate are predominantly present in the intestinal environment. A commensal bacterium, Bacteroides spp., mainly distributes succinate, which is subsequently consumed by secondary fermentative microbes in a steady state and therefore rarely accumulates in the intestinal environment. However, succinate is not consumed under antibiotic treatment or inflammatory conditions, eventually leading to its accumulation in the intestinal lumen. Succinate promotes gluconeogenesis of EHEC. In addition, the colonization and proliferation of C. rodentium are enhanced, especially with expression of virulence genes of the LEE. C. difficile can couple succinate metabolism and convert it to butyrate with the fermentation of carbohydrates, thereby enhancing its colonization and virulence. (E) Antibiotics can trigger the growth of Enterobacteriaceae. ROS at high concentrations result in an expansion of E. coli harboring an extra catalase that are genetically generated through chromosomal modification and eventually favor intestinal colonization of Vibrio cholerae, a strain that is highly sensitive strain to ROS, by reducing the ROS that are excessively generated in inflammatory conditions. E. coli, Escherichia coli; ROS, reactive oxygen species; RNS, reactive nitrogen species; iNOS, inducible nitric oxide synthase; EHEC, Enterohemorrhagic Escherichia coli; C. difficile, Clostridium difficile; C. rodentium, Citrobacter rodentium; SCFAs, short-chain fatty acids; LEE, locus of enterocyte effacement. |
 | Fig. 2Effects of antibiotics on the hypoxia barrier of intestinal epithelial cells. (A) In normal conditions, oxygen tension decreases steadily from the intestinal submucosal layer to the lumen. Although the partial pressure of oxygen is approximately 100 mm Hg in the basal layer, it is almost 0 mm Hg in the lumen. Under antibiotic treatment or inflammatory conditions, Clostridia produce butyrate and colonic epithelial cells convert the butyrate to carbon dioxide, leading to maintenance of hypoxia in the lumen. (B) When butyrate is lacking in the intestine, the cells utilize glucose for cellular respiration and the lactate that is released during the process increases oxygenation within the lumen. S. Typhimurium can proliferate using cytochrome bd-II oxidase encoded in cyxB, which is highly expressed at low oxygen concentrations. |
ACKNOWLEDGEMENTS
This research was supported by grants from the National Research Foundation (NRF) of Korea, funded by the Korean government (MSIP), 2017M3A9F3041233, 2017R1A2A2A05019987, 2015M3C9A2054024, and 2017R1A1A1A05001200.
References
1. Kumar H, Lund R, Laiho A, Lundelin K, Ley RE, Isolauri E, et al. Gut microbiota as an epigenetic regulator: pilot study based on whole-genome methylation analysis. MBio. 2014; 5:e02113–e02114.

2. Ha CW, Lam YY, Holmes AJ. Mechanistic links between gut microbial community dynamics, microbial functions and metabolic health. World J Gastroenterol. 2014; 20:16498–16517.

3. Yang BG, Hur KY, Lee MS. Alterations in gut microbiota and immunity by dietary fat. Yonsei Med J. 2017; 58:1083–1091.

4. Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017; 15:630–638.

7. Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, et al. The long-term stability of the human gut microbiota. Science. 2013; 341:1237439.

8. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012; 148:1258–1270.

9. Kamada N, Seo SU, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013; 13:321–335.

10. Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013; 14:685–690.

11. Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008; 3:213–223.

12. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012; 489:220–230.

13. Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med. 2016; 22:713–722.

14. Moya A, Ferrer M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 2016; 24:402–413.

15. Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol. 2011; 9:233–243.

16. Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008; 76:4726–4736.

17. Potgieter M, Bester J, Kell DB, Pretorius E. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol Rev. 2015; 39:567–591.

18. Allison SD, Martiny JB. Colloquium paper: resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci U S A. 2008; 105:Suppl 1. 11512–11519.
19. Becattini S, Taur Y, Pamer EG. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med. 2016; 22:458–478.

20. Pamer EG. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science. 2016; 352:535–538.

21. Comte J, Fauteux L, Del Giorgio PA. Links between metabolic plasticity and functional redundancy in freshwater bacterioplankton communities. Front Microbiol. 2013; 4:112.

22. Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest. 2014; 124:4212–4218.

23. Langdon A, Crook N, Dantas G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016; 8:39.

24. DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One. 2008; 3:e3056.

25. Bohnhoff M, Drake BL, Miller CP. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc Soc Exp Biol Med. 1954; 86:132–137.

26. Panda S, El khader I, Casellas F, López Vivancos J, García Cors M, Santiago A, et al. Short-term effect of antibiotics on human gut microbiota. PLoS One. 2014; 9:e95476.

27. Perez-Lopez A, Behnsen J, Nuccio SP, Raffatellu M. Mucosal immunity to pathogenic intestinal bacteria. Nat Rev Immunol. 2016; 16:135–148.

28. Schubert AM, Sinani H, Schloss PD. Antibiotic-induced alterations of the murine gut microbiota and subsequent effects on colonization resistance against Clostridium difficile. MBio. 2015; 6:e00974.

29. Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science. 2006; 313:89–92.

30. Gipponi M, Sciutto C, Accornero L, Bonassi S, Raso C, Vignolo C, et al. Assessing modifications of the intestinal bacterial flora in patients on long-term oral treatment with bacampicillin or amoxicillin: a random study. Chemioterapia. 1985; 4:214–217.
31. Nord CE, Sillerström E, Wahlund E. Effect of tigecycline on normal oropharyngeal and intestinal microflora. Antimicrob Agents Chemother. 2006; 50:3375–3380.

32. Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007; 1:56–66.

33. Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010; 5:e9836.

34. Gibson MK, Wang B, Ahmadi S, Burnham CA, Tarr PI, Warner BB, et al. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat Microbiol. 2016; 1:16024.

35. McFarland LV. Use of probiotics to correct dysbiosis of normal microbiota following disease or disruptive events: a systematic review. BMJ Open. 2014; 4:e005047.

36. Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008; 6:e280.

37. Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011; 108:Suppl 1. 4554–4561.

38. Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010; 3:148–158.

39. Pérez-Cobas AE, Gosalbes MJ, Friedrichs A, Knecht H, Artacho A, Eismann K, et al. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut. 2013; 62:1591–1601.

40. Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010; 8:292–300.

41. Spees AM, Wangdi T, Lopez CA, Kingsbury DD, Xavier MN, Winter SE, et al. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. MBio. 2013; 4:e00430–e00413.

42. Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013; 339:708–711.

43. Yoon MY, Min KB, Lee KM, Yoon Y, Kim Y, Oh YT, et al. A single gene of a commensal microbe affects host susceptibility to enteric infection. Nat Commun. 2016; 7:11606.

44. Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A. 2008; 105:2117–2122.

45. Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 2015; 17:662–671.

46. Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A. 2006; 103:10011–10016.

47. Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, et al. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol. 2006; 72:3593–3599.

48. Ley RE. Harnessing microbiota to kill a pathogen: the sweet tooth of Clostridium difficile. Nat Med. 2014; 20:248–249.

49. Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013; 502:96–99.

50. Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe. 2014; 16:770–777.

52. Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe. 2014; 16:759–769.

53. Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010; 467:426–429.

54. Rivera-Chávez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, et al. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. 2016; 19:443–454.

55. Lopez CA, Rivera-Chávez F, Byndloss MX, Bäumler AJ. The periplasmic nitrate reductase NapABC supports luminal growth of Salmonella enterica serovar Typhimurium during colitis. Infect Immun. 2015; 83:3470–3478.

56. El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013; 11:497–504.

57. Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011; 13:517–526.

58. Espey MG. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic Biol Med. 2013; 55:130–140.

59. Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015; 372:825–834.

60. Kelly CP, Pothoulakis C, LaMont JT. Clostridium difficile colitis. N Engl J Med. 1994; 330:257–262.

61. Johnston BC, Ma SS, Goldenberg JZ, Thorlund K, Vandvik PO, Loeb M, et al. Probiotics for the prevention of Clostridium difficile-associated diarrhea: a systematic review and meta-analysis. Ann Intern Med. 2012; 157:878–888.

62. Martz SL, McDonald JA, Sun J, Zhang YG, Gloor GB, Noordhof C, et al. Administration of defined microbiota is protective in a murine Salmonella infection model. Sci Rep. 2015; 5:16094.

63. Bakken JS, Borody T, Brandt LJ, Brill JV, Demarco DC, Franzos MA, et al. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol. 2011; 9:1044–1049.

64. Borody TJ, Khoruts A. Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol. 2011; 9:88–96.

65. Hassett DJ, Elkins JG, Ma JF, McDermott TR. Pseudomonas aeruginosa biofilm sensitivity to biocides: use of hydrogen peroxide as model antimicrobial agent for examining resistance mechanisms. Methods Enzymol. 1999; 310:599–608.
66. Schultz M. Clinical use of E. coli Nissle 1917 in inflammatory bowel disease. Inflamm Bowel Dis. 2008; 14:1012–1018.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download