Abstract
Purpose
microRNAs (miRNAs) are non-coding RNAs composed of 20 to 22 nucleotides that regulate development and differentiation in various organs by silencing specific RNAs and regulating gene expression. In the present study, we show that the microRNA (miR)-183 cluster is upregulated during hair cell regeneration and that its inhibition reduces hair cell regeneration following neomycin-induced ototoxicity in zebrafish.
Materials and Methods
miRNA expression patterns after neomycin exposure were analyzed using microarray chips. Quantitative polymerase chain reaction was performed to validate miR-183 cluster expression patterns following neomycin exposure (500 µM for 2 h). After injection of an antisense morpholino (MO) to miR-183 (MO-183) immediately after fertilization, hair cell regeneration after neomycin exposure in neuromast cells was evaluated by fluorescent staining (YO-PRO1). The MO-183 effect also was assessed in transgenic zebrafish larvae expressing green fluorescent protein (GFP) in inner ear hair cells.
Results
Microarray analysis clearly showed that the miR-183 cluster (miR-96, miR-182, and miR-183) was upregulated after neomycin treatment. We also confirmed upregulated expression of the miR-183 cluster during hair cell regeneration after neomycin-induced ototoxicity. miR-183 inhibition using MO-183 reduced hair cell regeneration in both wild-type and GFP transgenic zebrafish larvae.
Sensory-neural hearing loss (SNHL) is caused by aging, noise, and drugs and is one of the most common health problems associated with hearing loss. Although various treatment modalities have been developed, medical treatment for SNHL is still not available. The main SNHL pathology is cochlear hair cell damage, which is reversible in avians and fish. However, the mammalian cochlear hair cell is never regenerated, which is the main hurdle in medical treatment for SNHL. Therefore, understanding the hair cell regeneration mechanism in avians or fish is important to finding solutions for hair cell regeneration in mammals.
microRNAs (miRNAs), non-coding RNA composed of 20 to 22 nucleotides, regulate development and differentiation in various tissues by silencing specific RNA and regulating gene expression.1 miRNAs are expressed in the inner ear and have important roles in the development and maturation of the inner ear. For example, an miRNA (miR)-183 family cluster (miR-183, miR-96, and miR-182) is expressed in inner ear sensory cells in zebrafish2 and mice.3 Overexpression of miR-96 or miR-182 induces ectopic hair cell generation in zebrafish.4 In contrast, knockdown of miR-183, miR-96, and miR-182 causes reduced hair cell numbers in the inner ear.56 Moreover, mutations in miR-96 cause dominant-progressive hearing loss in mouse and humans.78
miRNAs have important roles during the regeneration of limbs, cardiac muscle, and liver.91011 Thus, an emerging question is whether miRNAs also control hair cell regeneration in the avian and fish inner ear. Previously, overexpression of miR-182, but not miR-183 and miR-96, was shown to improve hair cell survival after cisplatin treatment.12 Meanwhile, anti-miR-181a peptide reduced proliferation during hair cell regeneration after streptomycin exposure in avian inner ear. The let-7 family of miRNA is involved in the initial process of hair cell regeneration in newt inner ear.13 However, there has been no direct evi-dence supporting the role of miRNAs in hair cell regeneration.
Here, we show that miR-183 is essential for neuromast regeneration after neomycin-induced hair cell damage in zebrafish.
Wild-type adult zebrafish (AB) were obtained from a commercial supplier. The transgenic zebrafish lines that display green fluorescent protein (GFP) on hair cells (pou4f3::GFP) were also used.14 All procedures were following the regulation of the Institutional Animal Care and Use Committee of Yonsei University. Adult fish stock were maintained according to standard protocols.15 Embryos were raised at 28.5℃ in E3 embryo medium (1 mM MgSO4, 120 mM KH2PO4, 74 mM Na2HPO4, 1 mM CaCl2, 500 mM KCl, 15 mM NaCl, and 500 mM NaHCO3 in deionized H2O, pH 7.2) at a density of 30 larvae per 10-cm Petri dish.
Pulled-glass capillary micropipettes with tips broken to 10 µm were used to deliver 1 nL of morpholino (MO) into one-celled zebrafish embryos. The MO, including the standard control MO (GeneTools, Philomath, OR, USA), was dissolved in water to the desired concentration (1 mM) and injected into the cytoplasm. The sequence of anti-miR-183 (MO-183) was 5′-CAGTGAATCTACCAGTGCCATA-3′ & MO-182 was 5′-UUUGGCAAUGGUAGAACUCACA-3′. Wild-type zebrafish larvae at 72 hours post fertilization (hpf) were treated with 500 µM neomycin for 2 h. Hair cells from neomycin-treated larvae were stained using a 20-min immersion in 2 mM YO-PRO1 (Invitrogen, Cergy Pontoise, France) 4 h (78 hpf), 12 h (86 hpf), 24 h (98 hpf), and 48 h (122 hpf) following neomycin treatment. The zebrafish larvae were anesthetized with 0.1% tricaine (3-aminobenzoic acid ethyl ester, Sigma, St-Louis, MO, USA) and mo-unted in 3% methylcellulose (Sigma) in a depression slide. The average hair cell number in three posterior neuromasts (P7, P8, and P9) was evaluated under a fluorescent microscope. These posterior neuromasts were selected because they ex-hibit low variability in the number of hair cells, show definite differences in rate of regeneration, and produce clear confocal images due to the thin surrounding tissue. The study design is summarized in Fig. 1.
Zebrafish larvae were homogenized in Tri-reagent (Sigma) at a ratio of 100 mg of tissue per mL of reagent and total RNA was isolated at 4 h (78 hpf), 12 h (86 hpf), 24 h (98 hpf), and 48 h (122 hpf) following neomycin treatment. The miRNA expression profiles for each total RNA sample from zebrafish larvae were analyzed using GeneChip miRNA 1.0 arrays (Affymetrix, Santa Clara, CA, USA). Synthesis of complementary DNA (cDNA), hybridization to chips, and washes were performed according to the manufacturer's protocol. GeneChip arrays were scanned at 3-mm density using a GeneArray Scanner (Affymetrix). Images were inspected to ensure that all 18 chips had low background and bright hybridization signals. Mean fluorescence signal intensity for each probe was quantile normalized. The one-way ANOVA test was used to determine significant differences in miRNA expression between control groups, where p<0.05 was interpreted as significant.
Quantitative-polymerase chain reaction (q-PCR) detection of miRNAs was performed using mirVana q-PCR miRNA primer sets (Ambion, Cergy Pontoise, France). Total RNA (100 ng) from each group was reverse transcribed using SuperScript Reverse Transcriptase (Invitrogen) in a 20-mL reaction to produce cDNA. Q-PCR was performed using a 7500 Fast Real-Time PCR System (Applied Biosystems) to analyze triplicate reactions (20 mL) containing 26 SYBR Green PCR Master Mix (Applied Biosystems), a 10-fold dilution of the mirVana q-PCR Primer Set, and cDNA. After incubation at 95℃ for 20 s, PCR products were analyzed throughout 40 cycles consisting of 95℃ for 3 s and 60℃ for 30 s. The threshold cycle was defined as the PCR cycle number at which the fluorescence intensity was appreciably above the background level but was still in the early exponential phase of amplification.
RNA probes against miR-183 were hybridized to fish fixed at 120 hpf. Hybridization detection was via alkaline phosphatase (AP)-conjugated anti-digoxigenin (Roche, Indianapolis, IN, USA), followed by the NBT/BCIP color reaction. Whole-mount in situ hybridization was performed using locked nucleic acid (LNA) probes for miR-183 labeled with digoxigenin (DIG Oligonucleotide 3′-End Labeling Kit; Roche). RNA probes for in situ hybridization were synthesized using Taq polymerase. LNA probes (miRCURY LNA probes) were purchased from Exiqon (Vedbaek, Denmark) or custom synthesized (Integrated DNA Technologies, Coralville, IA, USA) by incorporating LNA modifications at every third nucleotide position from the 5′ end. LNA probes are antisense to miR-183 (5′-CAGTGAATTCTACCAGTGCCATA). Briefly, fixed tissues were defatted with ethanol, digested with 10 µg/mL Proteinase K/PBT, hybridized with 12 pmol labeled LNA probe, washed, and digested with RNase A. Labeled LNA probe was detected using AP conjugated sheep anti-DIG Fab fragment and BM Purple AP Substrate (Roche). Tissues were whole mounted in glycerol and imaged using light microscopy using a Nikon Eclipse 800 micro-scope. A minimum of three samples were prepared for each condition.
All experiments were performed independently at least three times using more than 10 larvae in each group for each experiment. Statistical analysis was performed using SPSS v.16.0 (SPSS Inc., Chicago, IL, USA). Tukey corrected one-way ANOVA or independent t-tests were used for analysis. Values of p<0.05 were considered statistically significant.
Microarrays were used to screen the miRNAs upregulated during hair cell regeneration after neomycin treatment. According to our previous data,15 hair cell regeneration starts 4 h after neomycin treatment and is completed within 24 h. Therefore, we compared miRNA expression levels at 4, 12, 24, and 48 h after neomycin treatment with those treated with vehicle. Fig. 2 lists the miRNAs with increases greater than log2-fold than control for at least one time point. Among the 37 miRNAs, we focused on the miR-183 cluster, which are expressed in hair cells and play important roles in hair cell development. miR-183 was upregulated 4 h after neomycin treatment, more st-rongly expressed at 12 h and 24 h after neomycin treatment, and then returned to basal levels 48 h after treatment. miR-182 and miR-96 had similar expression patterns. Other miRNAs, such as miR-21, miR-146a, and miR-739, were also increased more than log2 fold at three time points.
In order to validate the microarray data, we performed q-PCR for the miR-183 cluster. The miR-183 expression was significantly increased 12 h (3.3±0.7-fold) after neomycin treatment, further upregulated at 24 h (3.8±0.8-fold) after neomycin treatment, and returned to basal levels 48 h (1.2±0.1-fold) after neomycin treatment. miR-182 and miR-96 had similar expression patterns after neomycin treatment (Fig. 3).
In situ hybridization showed clear miR-183 expression in hair cells from zebrafish larvae (120 hpf) injected with control MO. In contrast, the signal was barely detected in larvae injected immediately after fertilization with MO-183 (1 mM) (Fig. 4). These experiments indicate that the MOs injected at fertiliza-tion work for at least 5 days. Next, we compared hair cell regeneration after neomycin treatment between MO-injected larvae and the control group. Hair cells labeled with YO-PRO-1 completely disappeared 4 h after treatment with 500 µM neomycin in lateral line neuromasts. In larvae injected with control MO, hair cell regeneration was evident 24 h after neomycin exposure. In contrast, hair cell regeneration was reduced in larvae injected with MO-183 (Fig. 5A and B). Fig. 5C shows the number of hair cells in the posterior neuromast (P7, P8, and P9) after neomycin treatment. The effects of MO-182 and MO-96 were similar to those of MO-183.
The effect of miR-183 knockdown on the regeneration of inner ear hair cells was also evaluated using zebrafish larvae expressing GFP in inner ear hair cells. The hair cells and otolith were completely regenerated 24 h after neomycin treatment (500 µM) in larvae injected with control MO (Fig. 6A). However, the hair cells were regenerated only partially in larvae injected with MO-183 (Fig. 6B).
Aberrant expression of miR-183 cluster has been identified in human disease, including cancer, neurological, and auto-immune disorders.1617 There are also several associations between miR-183 single nucleotide polymorphism and disease.18 Modulation of miR-183 expression has been shown to elicit therapeutic effects in animal disease models by regulating metabolism, apoptosis, and the immune system. Overexpression of miR-183 significantly inhibited the proliferation of nasopharyngeal cancer cells.19 miR-183 also have protective effect on ischemic liver injury.20 These results suggest that modulation of miR-183 cluster could be a potential therapeutic approaches for the treatment of deafness.
Previous research clearly shows that the miR-183 cluster plays an important role in hair cell development. Overexpression of miR-182 and miR-96 promotes zebrafish ectopic hair cell growth.4 Inhibition of the miR-183 cluster using antisense oligonucleotides results in delayed development of the zebrafish inner ear. Recently, evidence regarding the role of miRNAs in hair cell regeneration has been reported. Overexpression of miR-181a enhances the proliferation of chicken basilar papilla cells, which differentiate into myosin VI positive cells after streptomycin-induced hair cell loss.21 Conversely, inhibi-tion of miR-181a reduces papilla cells proliferation.22 These results indicate that miRNAs can be involved in hair cell regeneration as well as development. Here, we showed that the miR-183 cluster was upregulated in zebrafish larvae after neomycin-induced hair cell damage, and inhibition of miR-183 using antisense peptides resulted in reduced hair cell regeneration. According to previous reports, various miRNAs, including miR-183, are upregulated in the mouse inner ear following exposure to ototoxic drugs, noise stimulation, or oxidative st-ress.2324 Generation of oxidative stress during exposure to ototoxic drugs and noise stimulation can also induce hair cell degeneration.2526 Therefore, the miR-183 cluster may have a role in hair cell regeneration under stressful conditions in the mammalian inner ear, as well as in zebrafish.
Next, we examined how the miR-183 cluster stimulates hair cell regeneration in zebrafish. By combining several bioinformatics data sets, the miRNA targets involved in hair cell development were identified. In an ototoxicity mouse model induc-ed by kanamycin exposure, the inner ear exhibits an increase in the expression of miRNAs-34a and -34c.24 These miRNAs regulate the p53 gene, which regulates apoptosis. Similarly, various upregulated miRNAs after acoustic trauma also target genes that participate in regulating apoptosis. miR-183 also targets TAO kinase 1 and regulates cell death and apoptosis in rat cochlear cells.27 Inhibition of miR-183 reduces mature neural derivatives, such as astrocytes and neurons, by targeting SOX-2 in murine neural stem cell renewal.28 These data indicate that the miR-183 cluster can stimulate hair cell regeneration by regulating signal molecules involved in apoptosis or trans-differentiation of hair cells. Thus, the identification of exact miR-183 targets may provide new insight into the regulatory role of miR-183 during regeneration.
Our study design had a limitation regarding the lifespan of the MO. We injected the miR-183 morpholino immediately after fertilization. Although we confirmed that the expression of miR-183 was inhibited until 120 hpf, it is not clear if the miR-183 morpholino can completely inhibit miR-183 throughout this timeframe. Therefore, we are planning to evaluate the role of miRNAs in conditional tissue specific Dicer knockdown mice. Another on-going study is to assess the roles of miR-21, miR-146a, and miR-739, which also were upregulated in our microarray data, in hair cell regeneration.
In summary, we showed that the miR-183 cluster was upregulated during hair cell regeneration after ototoxic damage and that its inhibition reduced hair cell regeneration. Future studies to elucidate the exact targets and molecular mechanisms of miR-183 mediated hair cell generation will help to develop a new potential drug targeted to hearing loss.
Figures and Tables
 | Fig. 1Experimental design for the effect of microRNA in hair cell regeneration. MO-183 injection into zebrafish larvae immediately after fertilization. Larvae were treated with neomycin (500 µM) for 2 h at 72 hpf. Total RNA sampling and hair cell counting was performed at 4, 12, 24, and 48 h after neomycin treatment. hpf, hours post fertilization; MO, morpholino. |
 | Fig. 2miRNA expression microarray data after neomycin (500 µM) exposure for 2 h in zebrafish larvae. The data are expressed as log2 transformed fold change in miRNA expression at 4, 12, 24, and 48 h after neomycin treatment, compared to non-treated larvae. Only those miRNAs which showed 2- or more fold change at any time point, compared to control, are shown. Arrows indicated miRNAs up-regulated in three time points. *miR-183 clusters. miRNA, microRNA. |
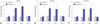 | Fig. 3Expression of microRNAs after neomycin exposure. miR-96 (A), miR-182 (B), and miR-183 (C) expression in zebrafish larvae after neomycin treatment was compared with that from non-treated samples (n=3 at each time point). *p<0.05. miR, miRNA. |
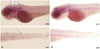 | Fig. 4In situ hybridization of miR-183 after MO injection. (A) The larvae injected with standard MO shows clear miR-183 expression in neuromast cells at 122 hours post fertilization (arrows). (B) In contrast, miR-183 expression disappeared in larvae injected with MO-183. Scale bar, 50 µm. miR, miRNA; MO, morpholino. |
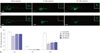 | Fig. 5The effect of miR-183 knockdown on hair cell regeneration after neomycin-induced ototoxicity. (A) Hair cells labeled with Y-PRO1 are regenerating 24 h after neomycin treatment in zebrafish larvae injected with control MO. (B) Hair cells are partially regenerating in larvae injected with MO-183 (1 mM). (C) Summarized data from six independent experiments. Scale bar, 100 µm. *p<0.05. miR, miRNA; MO, morpholino. |
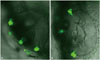 | Fig. 6The effect of miRNA-183 knockdown on hair cell regeneration in the inner ear of pou4f3::GFP zebrafish larvae. (A) Hair cells and otoliths are mostly regenerated in larvae injected with control MO. (B) Hair cells and otoliths are partially regenerated in larvae injected with MO-183. MO, morpholino; GFP, green fluorescent protein. |
ACKNOWLEDGEMENTS
This work was supported by the Myung Sun Kim Memorial Foundation (2015) to Jae Young Choi.
References
1. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297.
2. Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005; 309:310–311.

3. Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006; 1111:95–104.

4. Li H, Kloosterman W, Fekete DM. MicroRNA-183 family members regulate sensorineural fates in the inner ear. J Neurosci. 2010; 30:3254–3263.

5. Weston MD, Pierce ML, Jensen-Smith HC, Fritzsch B, Rocha-Sanchez S, Beisel KW, et al. MicroRNA-183 family expression in hair cell development and requirement of microRNAs for hair cell maintenance and survival. Dev Dyn. 2011; 240:808–819.

6. Friedman LM, Dror AA, Mor E, Tenne T, Toren G, Satoh T, et al. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci U S A. 2009; 106:7915–7920.

7. Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009; 41:614–618.

8. Mencía A, Modamio-Høybjør S, Redshaw N, Morín M, Mayo-Merino F, Olavarrieta L, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009; 41:609–613.

9. King BL, Yin VP. A conserved microRNA regulatory circuit is differentially controlled during limb/appendage regeneration. PLoS One. 2016; 11:e0157106.

11. Yi PS, Zhang M, Xu MQ. Role of microRNA in liver regeneration. Hepatobiliary Pancreat Dis Int. 2016; 15:141–146.

12. Li Y, Li A, Wu J, He Y, Yu H, Chai R, et al. MiR-182-5p protects inner ear hair cells from cisplatin-induced apoptosis by inhibiting FOXO3a. Cell Death Dis. 2016; 7:e2362.

13. Tsonis PA, Call MK, Grogg MW, Sartor MA, Taylor RR, Forge A, et al. MicroRNAs and regeneration: Let-7 members as potential regulators of dedifferentiation in lens and inner ear hair cell regeneration of the adult newt. Biochem Biophys Res Commun. 2007; 362:940–945.

14. Xiao T, Roeser T, Staub W, Baier H. A GFP-based genetic screen reveals mutations that disrupt the architecture of the zebrafish retinotectal projection. Development. 2005; 132:2955–2967.

15. Moon IS, So JH, Jung YM, Lee WS, Kim EY, Choi JH, et al. Fucoidan promotes mechanosensory hair cell regeneration following amino glycoside-induced cell death. Hear Res. 2011; 282:236–242.

16. Dai R, Zhang Y, Khan D, Heid B, Caudell D, Crasta O, et al. Identification of a common lupus disease-associated microRNA expression pattern in three different murine models of lupus. PLoS One. 2010; 5:e14302.

17. Yu H, Liu Y, Bai L, Kijlstra A, Yang P. Predisposition to Behçet's disease and VKH syndrome by genetic variants of miR-182. J Mol Med (Berl). 2014; 92:961–967.

18. Sánchez-Mora C, Ramos-Quiroga JA, Garcia-Martínez I, Fernàndez-Castillo N, Bosch R, Richarte V, et al. Evaluation of single nucleotide polymorphisms in the miR-183-96-182 cluster in adulthood attention-deficit and hyperactivity disorder (ADHD) and substance use disorders (SUDs). Eur Neuropsychopharmacol. 2013; 23:1463–1473.

19. Wang G, Wang S, Li C. MiR-183 overexpression inhibits tumorigenesis and enhances DDP-induced cytotoxicity by targeting MTA1 in nasopharyngeal carcinoma. Tumour Biol. 2017; 39:1010428317703825.

20. Lin HC, Liu SY, Yen EY, Li TK, Lai IR. microRNA-183 mediates protective postconditioning of the liver by repressing Apaf-1. Antioxid Redox Signal. 2017; 26:583–597.

21. Frucht CS, Uduman M, Duke JL, Kleinstein SH, Santos-Sacchi J, Navaratnam DS. Gene expression analysis of forskolin treated basilar papillae identifies microRNA181a as a mediator of proliferation. PLoS One. 2010; 5:e11502.

22. Frucht CS, Santos-Sacchi J, Navaratnam DS. MicroRNA181a plays a key role in hair cell regeneration in the avian auditory epithelium. Neurosci Lett. 2011; 493:44–48.

23. Wang Z, Liu Y, Han N, Chen X, Yu W, Zhang W, et al. Profiles of oxidative stress-related microRNA and mRNA expression in auditory cells. Brain Res. 2010; 1346:14–25.

24. Yu L, Tang H, Jiang XH, Tsang LL, Chung YW, Chan HC. Involvement of calpain-I and microRNA34 in kanamycin-induced apoptosis of inner ear cells. Cell Biol Int. 2010; 34:1219–1225.

25. Henderson D, Bielefeld EC, Harris KC, Hu BH. The role of oxidative stress in noise-induced hearing loss. Ear Hear. 2006; 27:1–19.

26. Jiang H, Talaska AE, Schacht J, Sha SH. Oxidative imbalance in the aging inner ear. Neurobiol Aging. 2007; 28:1605–1612.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download