Abstract
Only 6 patients with partial trisomy of the long arm of chromosome 19 (19q), caused by direct interstitial duplications, have been reported until today. Herein, we report a pediatric patient with a novel 1.13 Mb direct interstitial duplication within 19q13.32, which is the smallest fragment affected so far. A five-year old Korean boy of healthy parents presented with microcephaly, growth retardation, developmental delay, and craniofacial dysmorphism. Even though G-banded chromosome analysis at resolution of 550-band revealed normal karyotype, duplication of 1.13 Mb fragment within 19q13.32 was detected by array comparative genomic hybridization. Comparing with previously reported patients with pure duplication involving 19q as a sole chromosomal abnormality, our case showed the smallest duplication segment with relatively mild degree of clinical features. Our present case might serve as the landmark case among patients with 19q duplication for genotype-phenotype correlation study and further identification of critical region for 19q duplication abnormalities.
A few patients with partial trisomy of the long arm of chromosome 19 (19q), caused by direct interstitial duplications, have been reported until today.1 Although there were several cases of partial trisomy involving 19q with other chromosomal rearrangements as a complex karyotype,2 only 6 patients with pure duplication, involving 19q as a sole chromosomal abnormality, have been reported with several common phenotypes such as developmental delay and dysmorphic features.345678 In these cases, different loci ranging from 19q11.05 to 19q13.4 were duplicated with various sizes of affected segment. Herein, we describe a new case of a five-year old Asian boy with a novel 1.13 Mb direct interstitial duplication within 19q13.32 region, which is the smallest fragment affected compared to previous case reports with pure duplication involving 19q.
The proband, a Korean male, was born at 39 weeks of gestation by cesarean section due to cephalopelvic disproportion as the first son of the parents. The mother was 29 years old and the father was 29 years old, who were both healthy and had no remarkable family history. His birth weight, length, and head circumference were 2500 g (3 percentile), 46 cm (10 percentile), and 33 cm (25 percentile), respectively. Even though microce-phaly was concerned for medical follow up, the physical examination at birth revealed no specific abnormality. When the patient was 8-month old, microcephaly was recognized with the head circumference of 42 cm (3 percentile). Delayed achievement for developmental milestones was observed when he was 12-month old: he could start independent walking at 14 months, could call mom at 33 months, and bladder training was possible at 28 months. Bailey scales of infant development, performed when he was 38-month old, revealed the retardation in the following parts; fine motor delayed for 11 months, gross motor and social function delayed for 14 months, and cognition and speech delayed for 22 months. Limited growth rate in terms of weight gain was also noticed (i.e., 16 kg at the age of 5 years). On physical examination, craniofacial dysmorphic features such as broad nasal bridge, high forehead with posterior hairline, and upward directed corners of the eyes were noticed but the degree of dysmorphism was relatively mild. Additionally, mild degree of pes planovalgus was present.
Chromosomal analysis performed with peripheral blood by conventional G-banding technique at the 550-band resolution revealed the normal karyotype, 46,XY[20] (Fig. 1A). However, when array comparative genomic hybridization (CGH) was performed with NimbleGen CGX-3 135K whole-genome array (Roche NimbleGen, Inc., Madison, WI, USA) by the commercially available Genoglyphix software (Signature Genomics, Spokane, WA, USA), a 1.13 Mb (chr19:51839641–52967920) duplication within 19q13.32 region was detected (Fig. 1B). Chromosomal analysis and array CGH studies on both parents revealed no abnormal findings, indicating that the 1.13 Mb duplication found in the patient was a de novo rearrangement.
During the follow-up outpatient clinics, he did not show any additional symptom or sign associated with neurologic dysfunction and cardiac problem. The brain MRI showed no abnormal findings for pathologic lesion. Until the day of publication, the patient was treated with supportive care and tolerated well in the public childcare facility.
The scarcity of pure interstitial duplication of 19q has impeded the classification and identification of “19q duplication syndrome,” based on the common phenotypes, in the previous case reports. Wide spectrum and variable expressivity of clinical features are recognized in several categories; developmental delay, craniofacial dysmorphic findings, growth abnormality presented as obesity or growth retardation, skeletal defects, and brain anomaly or seizures (Table 1). Interestingly, this is the first Asian report among patients with 19q duplication. In three previous cases with prenatally diagnosed duplication of 19q, the authors did not describe phenotypic findings.91011 When we compared the present case with 6 previously reported 19q duplication patients, this patient showed relatively mild manifestations (Table 1). Our patient might be highlighted for the smallest size of duplication fragment involved among patients with 19q duplication until today. Two important clinical characteristics which were commonly present in our case and other patients are developmental delay and microcephaly. Therefore, we think that the candidate genes fundamental for these two phenotypes could be narrowed to the genes within the 19q13.32 region, the 1.13 Mb (chr19:47147801–48276108 by GRCh37/hg19; chr19:51839641–52967920 by NCBI36/hg18) segment.
Davidsson, et al.12 recently studied the association between obesity and dup(19)(q12q13.2), and suggested several candidate genes within the region from 19q12 to 19q13.2, such as AKT2, CEACAM1, CEBPA, and TGFB1, in context of probable new obesity-related syndrome. Although their conclusions on the association between obesity and patients carrying dup(19) (q12q13.2) were equivocal, similar genotype-phenotype correlation approach is needed for patients with pure direct interstitial 19q duplication.
Interestingly, reciprocal deletions which involved the analogous region of 19q were reported in some cases.1314 Most importantly, the recent case reports which presented the smallest overlapping fragment of deletion established the definition of novel 19q13.11 deletion syndrome with identification of the critical region.15 From this point, our case might be highlighted for the specific region of duplication in 19q.
Twenty-five protein-coding genes are located within the duplicated region, among which only 3 genes are currently known with specific diseases; FKRP, AP2S1, and KPTN. Although these genes have been suggested to be associated with muscular dystrophy, hypercalcemia, and mental retardation, respectively, no such distinct phenotype was observed in our patient, possibly because it was duplication rather than deletion. When we compared 5 patients from DECIPHER database with duplicated segments within the affected region of our patient, 80% and 40% presented developmental delay and microcephaly, respectively. Even though duplicated segment sizes in these 5 patients ranged from 38 kb to 897 kb, it was not possible to conclusively determine specific genes responsible for clinical features of microcephaly and developmental delay. Although further genetic test using whole exome sequencing might discover additional variants, we conclude that 1.13 Mb interstitial duplication at 19q13.32 might have greatly contributed to cardinal phenotypes in this patient.
When we investigated diverse chromosome loci which are associated with the phenotype of developmental delay, deletion rather than duplication appeared to be predominantly related with the developmental delay,1617 thus leading to a possibility that the gain of genetic content due to duplication might lead to excessive formation of material that would hinder the normal development. It is also likely that clinical manifestations which were not observed in our case, such as cardiac malformation and seizure, might be caused by specific genes located outside the 19q13.32 region and within the 19q1 band. However, it would be necessary to remember that late-onset symptoms or signs can be expressed as the pediatric patient develops.
In conclusion, we report the first Asian patient with de novo 1.13 Mb interstitial duplication within 19q13.32 region, who presented developmental delay and microcephaly. It should be noted that our patient might serve as the landmark case for genotype-phenotype correlation study in 19q duplication patients.
Figures and Tables
Fig. 1
(A) Karyotype analysis. Trypsin-Giemsa banded chromosome analysis at the 550-band level shows a normal karyotype, 46,XY[20]. (B) Genome analysis using the array-comparative genomic hybridization. Duplication within 19q13.32 region is observed. The size of duplication fragment was estimated to be 1.13 Mb (chr19:47147801-48276108). Duplicated segments and clinical features of 5 patients from DECIPHER database with duplicated segments within the affected region of our patient are comparatively presented below.
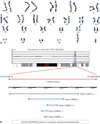
Table 1
Summary of Clinical and Genetic Features Observed in Live-Born Cases of Pure Duplication Involving 19q

| Present patient | Quack, et al.3 | Bhat, et al.4 | Qorri, et al.5 | Zung, et al.6 | Palomares, et al.7 | Lugli, et al.8 | |
|---|---|---|---|---|---|---|---|
| Sex | Male | Male | Male | Female | Male | Female | Male |
| Age | 5 years | 48 months | 18 months | 27 months | 14 years | 15 months | 36 months |
| Race | Asian | Caucasian | Caucasian | Caucasian | NA | Caucasian | Caucasian |
| Karyotype | 46,XY* | 47,XY,r(19) (q11.05q13.2) | 46,XY,dup(19) (q13.3q13.4) | 46,XX,dup(19) (q13.1q13.3) | 47,XY,+der(19) (q12q13.2) | 46,XX,dup(19) (q12q13.2) | 46,XY,dup(19) (q12q13.2) |
| Duplicated segment (size) | 19q13.32 (1.13 Mb) | 19q11.05–13.2 (NA) | 19q13.3–13.4 (NA) | 19q13.1–13.3 (NA) | 19q12–13.2 (NA) | 19q12–13.2 (10.8 Mb) | 19q12–13.2 (12.4 Mb) |
| Genetic diagnosis tools | Array CGH | FISH | FISH | FISH | FISH and array CGH | FISH and array CGH | FISH and array CGH |
| Clinical features | |||||||
| Developmental delay | + | + | + | + | + | + | + |
| Growth retardation or obesity | Growth retardation | Obesity | Growth retardation | − | Obesity | − | − |
| Microcephaly or macrocephaly | Microcephaly | Macrocephaly | Microcephaly | − | Macrocephaly | Microcephaly | − |
| Craniofacial dysmorphism | + | + | + | − | + | + | + |
| Abnormal ear including strabismus | − | − | + | − | + | + | + |
| Skeletal defect | + | − | + | − | − | − | − |
| Brain anomaly or seizure | − | − | − | − | + | + | + |
| Cardiac malformation | − | − | + | − | − | − | + |
ACKNOWLEDGEMENTS
The authors thank the parents for agreeing to participate in this study. We thank also all the technical team for array-CGH experiments.
References
1. Boyd E, Grass FS, Parke JC, Knutson K, Stevenson RE. Duplication of distal 19q: clinical report and review. Am J Med Genet. 1992; 42:326–330.

2. Sauter SM, Böhm D, Bartels I, Burfeind P, Laccone FA, Neesen J, et al. Partial trisomy of distal 19q detected by quantitative realtime PCR and FISH in a girl with mild facial dysmorphism, hypotonia and developmental delay. Am J Med Genet A. 2007; 143A:1091–1099.

3. Quack B, Van Roy N, Verschraegen-Spae MR, Klein F. Interstitial deletion and ring chromosome derived from 19q. Proximal 19q trisomy phenotype. Ann Genet. 1992; 35:241–244.
4. Bhat M, Morrison PJ, Getty A, McManus D, Tubman R, Nevin NC. First clinical case of small de novo duplication of 19q (13.3-13.4) confirmed by FISH. Am J Med Genet. 2000; 91:201–203.

5. Qorri M, Oei P, Dockery H, McGaughran J. A rare case of a de novo dup(19q) associated with a mild phenotype. J Med Genet. 2002; 39:E61.

6. Zung A, Rienstein S, Rosensaft J, Aviram-Goldring A, Zadik Z. Proximal 19q trisomy: a new syndrome of morbid obesity and mental retardation. Horm Res. 2007; 67:105–110.

7. Palomares Bralo M, Delicado A, Lapunzina P, Velázquez Fragua R, Villa O, Angeles Mori M, et al. Direct tandem duplication in chromosome 19q characterized by array CGH. Eur J Med Genet. 2008; 51:257–263.

8. Lugli L, Malacarne M, Cavani S, Pierluigi M, Ferrari F, Percesepe A. A 12.4 Mb direct duplication in 19q12-q13 in a boy with cardiac and CNS malformations and developmental delay. J Appl Genet. 2011; 52:335–339.

9. Cotter PD, McCurdy LD, Gershin IF, Babu A, Willner JP, Desnick RJ. Prenatal detection and molecular characterization of a de novo duplication of the distal long arm of chromosome 19. Am J Med Genet. 1997; 71:325–328.

10. Tercanli S, Hösli I, Berlinger A, Beyer R, Achermann J, Holzgreve W. Prenatal diagnosis of a partial trisomy 19q. Prenat Diagn. 2000; 20:663–665.

11. Babic´ I, Brajenovic´-Milic´ B, Petrovic´ O, Mustac´ E, Kapovic´ M. Prenatal diagnosis of complete trisomy 19q. Prenat Diagn. 2007; 27:644–647.

12. Davidsson J, Jahnke K, Forsgren M, Collin A, Soller M. dup(19) (q12q13.2): array-based genotype-phenotype correlation of a new possibly obesity-related syndrome. Obesity (Silver Spring). 2010; 18:580–587.

13. Kulharya AS, Michaelis RC, Norris KS, Taylor HA, Garcia-Heras J. Constitutional del(19)(q12q13.1) in a three-year-old girl with severe phenotypic abnormalities affecting multiple organ systems. Am J Med Genet. 1998; 77:391–394.

14. Malan V, Raoul O, Firth HV, Royer G, Turleau C, Bernheim A, et al. 19q13.11 deletion syndrome: a novel clinically recognisable genetic condition identified by array comparative genomic hybridisation. J Med Genet. 2009; 46:635–640.

15. Schuurs-Hoeijmakers JH, Vermeer S, van Bon BW, Pfundt R, Marcelis C, de Brouwer AP, et al. Refining the critical region of the novel 19q13.11 microdeletion syndrome to 750 Kb. J Med Genet. 2009; 46:421–423.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download