Abstract
Purpose
This study was conducted to identify and to functionally characterize genetic variants in ST3GAL5 and ST8SIA1 in Korean patients with thyroid-associated ophthalmopathy (TAO).
Materials and Methods
Genetic analyses were conducted using DNA samples from TAO patients (n=50) and healthy subjects (n=48) to identify TAO-specific genetic variants of ST3GAL5 or ST8SIA1. The effect of each genetic variant on the transcription or expression of these genes was examined. Additionally, correlations between functional haplotypes of ST3GAL5 or ST8SIA1 and clinical characteristics of the patients were investigated.
Results
Six promoter variants and one nonsynonymous variant of ST3GAL5 were identified, and four major promoter haplotypes were assembled. Additionally, three promoter variants and two major haplotypes of ST8SIA1 were identified. All ST3GAL5 and ST8SIA1 variants identified in TAO patients were also found in healthy controls. Promoter activity was significantly decreased in three promoter haplotypes of ST3GAL5 and increased in one promoter haplotype of ST8SIA1. Transcription factors activating protein-1, NKX3.1, and specificity protein 1 were revealed as having roles in transcriptional regulation of these haplotypes. The nonsynonymous variant of ST3GAL5, H104R, did not alter the expression of ST3GAL5. While no differences in clinical characteristics were detected in patients possessing the functional promoter haplotypes of ST3GAL5, exophthalmic values were significantly lower in patients with the ST8SIA1 haplotype, which showed a significant increase in promoter activity.
Thyroid-associated ophthalmopathy (TAO) is an ophthalmic disease that frequently accompanies Graves' disease (GD). In general, 25–50% of patients with GD are diagnosed with clinically significant TAO.1 TAO follows a characteristic clinical course, called Rundle's curve,2 in which TAO worsens during an initial phase, peaks at maximum severity, improves, and reaches a static plateau, with the activity curve preceding the severity curve by a few months. Throughout the course of the disease, patients present with characteristic symptoms of lid swelling, chemosis, proptosis, lid retraction, diplopia due to limited ocular movement, and deep orbital pain, each with variable severity. While most patients experience remission through cessation of the active inflammatory stage by medical interferences, such as administration of corticosteroids, without a need for additional treatment, a considerable number of patients require surgical interventions, such as orbital decompression for proptosis, strabismus surgery, and levator recession for lid retraction. Moreover, approximately 3–5% of patients with TAO suffer from severe disease with intense inflammation, resulting in devastating complications, such as sight-threatening corneal ulceration and compressive optic neuropathy.3
Smoking is widely accepted as a critical environmental risk factor that affects the severity of TAO.4 Smokers with TAO are more likely than nonsmokers to develop severe disease and less likely to respond well to immunosuppressive therapies,5 which provides concrete evidence for discouraging smoking in TAO patients. With the development of technologies for genetic study, an increasing number of researchers have investigated the association between genetic variants and susceptibility to or severity of numerous diseases, including TAO. Polymorphisms in several genes including those encoding interleukin (IL)-1β, IL-12B, toll-like receptors, poly (ADP-ribose) polymerase-1, and nuclear factor-kappa B (NF-κB) are significantly associated with TAO susceptibility.6789
ST3GAL5 (lactosylceramide alpha-2,3-sialyltransferase) and ST8SIA1 (ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1) are members of the sialyltransferase family and are involved in the production of gangliosides. By enzymatic activity that transfers sialic acid to nascent oligosaccharide, ST3GAL5 converts lactosylceramide into monosialodihexosylganglioside 3 (GM3), and ST8SIA1 converts GM3 to disialoganglioside 3 (GD3), which is a precursor of trisialoganglioside 1b (GT1b).10 Previously, we observed that mRNA levels of ST3GAL5 and ST8SIA1 increased in orbital fat tissues obtained from TAO patients, compared with those from healthy controls.11 In addition, GT1b induced an increase in hyaluronic acid (HA) in orbital fibroblasts.11 HA is a key molecule in determining TAO severity, based on its hydrophilic characteristics and its ability to induce tissue edema by accumulation in orbital connective tissue and extraocular muscles.12 Taken together, one can postulate a possible impact of ST3GAL5 or ST8SIA1 genetic variants and the resulting alteration of their activity on susceptibility to and/or severity of TAO. However, to the best of our kn-owledge, a genetic variant of ST3GAL5 or ST8SIA1 has not yet been studied in TAO patients.
In the present study, we conducted genetic analyses using DNA samples from 50 TAO patients to identify the TAO-specific genetic variants of ST3GAL5 or ST8SIA1. In addition, we examined the effect of each genetic variant on the transcription or expression of these genes. Finally, we investigated correlations between functional haplotypes of ST3GAL5 and ST8SIA1 and the clinical characteristics of the patients.
This study was approved by the Institutional Review Board of Ajou Medical Center, Suwon, Korea and the Institutional Review Board of Severance Hospital in the Yonsei University Health System, Seoul, Korea. All experiments and analyses were performed in accordance with relevant guidelines and regulations of the Institutional Review Boards of Ajou Medical Center and the Yonsei University Health System. DNA samples were obtained from peripheral blood upon obtaining written informed consent from 50 Korean patients with TAO (8 males and 42 females); those diagnosed with TAO had at least a 12-month follow-up period prior to the enrollment in this study. Mean age was 42.1±14.0, and the follow-up period ranged from 12 to 78 (28.6±19.4) months. During this period, the patient assessments were performed by a single oculoplastic specialist in each hospital. According to the No signs or symptoms, Only signs without symptoms, Soft tissue involvement, Proptosis, Extraocular muscle involvement, Corneal involvement, Sight loss (NOSPECS) classification,13 patients with dysthyroid disease, showing symptoms in addition to soft tissue involvement, were diagnosed with TAO. Clinical activity was measured by the clinical activity score (CAS), which included clinical symptoms and signs of TAO (Table 1).14 For the exophthalmos measurements, the same type of Hertel exophthalmometer was used at both hospitals, which has been shown to be a reliable method for measuring ocular protrusion, showing good intra-observer agreement and clinically acceptable inter-observer agreement without significant variations.15 Clinical data were collected by reviewing medical records. For the control group, DNA samples from 48 healthy Koreans were obtained from DNA Link, Inc. (Seoul, Korea), a genomic service provider established in March 2000 and based in Korea focusing on research and development in the field of bio-industry and genomic analysis industry.
Genetic variants of ST3GAL5 and ST8SIA1 were identified by direct sequencing of the promoter (-2295 through +417, and -2539 through +176, from translation start site, respectively) and whole exons. Haplotype assembly was conducted using Haploview software, version 4.3 (Broad Institute, Cambridge, MA, USA). The GenBank mRNA sequences (accession numbers NM_003896 and NM_003034) were used as reference sequences for ST3GAL5 and ST8SIA1, respectively, in this study.
To construct the ST3GAL5 reporter plasmid, 2040-bp regions of the gene were amplified and inserted into the pGL4.11b[luc2] vector (Promega Corporation, Fitchburg, WI, USA). The ST8SIA1 reporter plasmid was constructed by amplifying and inserting 1977-bp regions into the pGL4.11[luc2P] vector (Promega Corporation). To evaluate the effect of variants in the coding region on ST3GAL5 expression, we constructed the ST3GAL5 reference plasmid; ST3GAL5 cDNA was purchased (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) and subcloned into the pcDNA3.1(+) vector (Life Technologies Corporation, Carlsbad, CA, USA). Haplotypes or variants in the promoter region and a nonsynonymous variant were generated using a QuikChange2® II site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). All DNA sequences were confirmed by direct sequencing. Supplementary Table 1 (only online) lists the primers used in this study.
Previously, we measured luciferase activities of several genes after transfection of reporter vectors into different cell line, such as HCT-116, HEK-293, HeLa, ACHN, and HepG2.161718 Then we observed that, regardless of the type of overexpressed gene, HCT-116 (human colon carcinoma) cells showed the highest promoter activity. Therefore, after transfection of ST3GAL5 and ST8SIA1 reporter plasmids into HCT-116 cells using Lipofectamine LTX and Plus reagents (Life Technologies Corporation), the luciferase activity of each vector was measured using a Dual-Luciferase2® Reporter Assay System (Promega Corporation), according to the manufacturer's protocol. A Renilla vector was used to equalize the amounts of each transfected plasmid.
TFSearch (version 3.1, Real-World Company Partnership, Tokyo, Japan), MatInspector (Genomatix Software GmbH, Munich, Germany), and ConSite (http://consite.genereg.net/cgi-bin/consite) were used to predict transcription factors that bind to the ST3GAL5 or ST8SIA1 promoter regions with binding affinities that would be affected by their variants. For electrophoretic mobility shift assay (EMSA), nuclear proteins from HCT-116 cells were incubated with 32P-labeled oligonucleotides for 30 min at room temperature. Protein-DNA mixtures were then separated by 6% non-denaturing polyacrylamide gel electrophoresis. For the competition assay, 50- or 100-fold molar excess of unlabeled consensus or core sequence-mutated oligonucleotides was used. In the EMSAs for activating protein-1 (AP-1) and specificity protein 1 (SP1), supershift assays were performed using 4 µg of antibodies against phospho-c-Jun (9164, Cell Signaling Technology, Danvers, MA, USA) and SP1 (sc-420X, Santa Cruz Biotechnology, CA, USA), respectively. The oligonucleotides used in the EMSA are listed in Supplementary Table 2 (only online).
The ST3GAL5 reference or variant plasmids were transfected into HCT-116 cells. Forty-eight hours after transfection, cell lysates were obtained, and immunoblotting was carried out. A rabbit anti-ST3GAL5 antibody (Abcam, Cambridge, UK) was used as the primary antibody, and β-actin was used as an internal standard.
Data analysis was conducted using IBM SPSS Statistics software, version 21 (IBM Corp., Armonk, NY, USA). Luciferase activity or protein expression was compared between the ST3GAL5 or ST8SIA1 references and their variants by one-way analysis of variance, followed by a Dunnett's two-tailed test. To examine the effect of ST3GAL5 or ST8SIA1 functional haplotypes on the clinical course of TAO, we used the Mann–Whitney nonparametric test. Data are represented as mean values±standard deviation (SD). p-values of less than 0.05 were considered statistically significant.
Sequencing of genomic DNA samples from 50 TAO patients revealed six genetic variants in the promoter region of ST3GAL5 and one nonsynonymous variant. In the case of ST8SIA1, three promoter variants were identified, while there was no nonsynonymous variant of this gene in our population. Table 2 shows the frequencies of genetic variants of ST3GAL5 and ST8SIA1. However, we found that those variants were not TAO-specific: all variants identified in TAO patients were observed in the 48 healthy controls, and the frequency of each variant was comparable between the two groups (Supplementary Table 3, only online). After haplotype construction with the genotype data, we observed four and two common (frequency ≥5%) promoter haplotypes in ST3GAL5 and ST8SIA1, respectively (Tables 3 and 4). Of those haplotypes, haplotype 2 (H2) of ST3GAL5 and haplotype 1 (H1) of ST8SIA1 were used as reference haplotypes in the present study, based on accession numbers NM_003896 and NM_003034 in the National Center for Biotechnology Information database of single-nucleotide polymorphisms (SNPs).
To the best of our knowledge, no studies have investigated the effect of ST3GAL5 or ST8SIA1 promoter variants on the transcription of these genes. Here, we constructed vectors containing ST3GAL5 and ST8SIA1 promoter variants or haplotypes and measured the luciferase activity of each vector. Three ST3GAL5 haplotypes exhibited a significant decrease in promoter activity, compared with the reference (H2) (Fig. 1A); promoter activities of H1, H3, and H4 were decreased by 13.45, 39.04, and 17.32%, respectively. Two variants, g.-1468C>G and g.-1051G>C, present in H1, H3, and H4, exhibited significantly decreased promoter activities, compared with the reference, by 33.53 and 39.0%, respectively (Fig. 1B). In the case of ST8SIA1, H2 showed increased promoter activity, compared with the reference (H1) (Fig. 1C), by 102.30%. Two variants, g.-2196A>C and g.-1984T>C, in H2 were involved in an increase in activity of H2; they showed increased promoter activities, compared with the reference, by 40.37 and 70.36%, respectively (Fig. 1D).
We then conducted transcription factor binding site (TFBS) analyses to predict which transcription factors would bind to the ST3GAL5 or ST8SIA1 promoter regions and were involved in the transcriptional regulation of genetic variants. TFSearch and MatInspector predicted that the transcription factors AP-1 and v-Myb would bind in the g.-1051G>C and g.-1468C>G variant regions, respectively, and that the binding affinities of these transcription factors would be affected by each genetic variant. To validate the TFBS, we performed EMSAs. To examine AP-1 binding to the g.-1051G>C variant, nuclear extracts were incubated with 32P-labelled oligonucleotides (lanes 1–3, AP-1 consensus; lanes 4–6, g.-1051G reference; lanes 7–9, g.-1051C variant) (Fig. 2A), and the reaction mixtures were separated by electrophoresis. We observed that reference or variant oligonucleotides formed DNA-protein complexes at the same position as the AP-1 consensus oligonucleotide-protein complex; however, the intensity of the variant oligonucleotides-protein complex increased by 12%, compared with the reference (lanes 1, 4, and 7) (Fig. 2A). The competition assay using a 100-fold molar excess of unlabeled AP-1 consensus oligonucleotides
revealed that the DNA-protein complexes contained AP-1 (lanes 2, 5, and 8) (Fig. 2A). To validate the competition assay, we performed a supershift assay using an antibody against AP-1 (lanes 3, 6, and 9) (Fig. 2A). The DNA-protein complexes supershifted with the antibody, indicating that these bands contained complexes of DNA with AP-1. To determine whether v-Myb could bind to the g.-1468C>G variant region, we performed a gel shift assay and confirmed that v-Myb bound to the ST3GAL5 g.-1468C>G promoter region. However, the binding affinity for v-Myb was comparable between the reference and variant (Supplementary Fig. 1, only online). In the case of ST8SIA1, two transcription factors, NKX3.1 and SP1, were predicted to bind in the g.-2196A>C and g.-1984T>C> variant regions of ST8SIA1, respectively. We observed that NKX3.1 had a higher affinity to the reference g.-2196A than the variant g.-2196C (lanes 4 and 7) (Fig. 2B) through a competition assay using a 100-fold molar excess of unlabeled NKX3.1 consensus (lanes 5 and 8) (Fig. 2B) or core sequence-mutated oligonucleotides (lanes 6 and 9) (Fig. 2B). Finally, in the competition assay using a 100-fold molar excess of unlabeled SP1 consensus oligonucleotides (lanes 5 and 8) (Fig. 2C), SP1 bound to the promoter region near the g.-1984T>C variant, and the binding affinity was increased by the presence of this variant (lanes 4 and 7) (Fig. 2C). The supershift using the antibody against SP1 confirmed that these DNA-protein complexes contained SP1 (lanes 3, 6, and 9) (Fig. 2C).
In general, nonsynonymous variants resulting in amino acid changes frequently alter the expression or function of genes. In the present study, we identified one nonsynonymous variant of ST3GAL5, H104R. To date, the effect of this variant on ST3GAL5 expression is not known. Therefore, we constructed a vector containing ST3GAL5 reference sequences or the H104R variant, and used immunoblotting to determine whether H104R affects ST3GAL5 expression. We observed that ST3GAL5 expression with the H104R variant was similar to that with the reference (Fig. 3).
Although genetic variants or haplotypes of ST3GAL5 and ST8SIA1 identified in this study are not TAO-specific, we investigated whether functional haplotypes of these genes affect the clinical severity thereof during patient follow-up. To examine this, the demographic and clinical characteristics of patients, including CAS, exophthalmic value, number of cyclic treatments with steroids, and total amount of steroid used, were collected and compared according to the ST3GAL5 or ST8SIA1 functional promoter haplotypes. Of the 50 patients, 15 were excluded from these comparisons owing to missing data. The ST3GAL5 functional promoter haplotypes H1, H3, and H4 did not affect the clinical characteristics of TAO patients (Table 5). However, we observed that patients with ST8SIA1 H2 had a lower exophthalmic value at the first visit, compared with patients without H2 (19.11±1.58 mm in the variant vs. 20.99±3.52 mm in the control groups, p=0.007). In addition, the peak exophthalmic value of patients with H2 during the follow-up period was less than that in those without H2 (19.75±2.02 mm in the variant vs. 22.00±3.42 mm in the control groups, p=0.003) (Table 6). Other parameters, including CAS at the first visit, peak CAS during the follow-up period, total amount of steroid used, and number of steroid pulse cycles, were comparable between the two groups. Demographic characteristics, including age, gender, length of follow-up period, family history, smoking status, and comorbidities (hypertension and diabetes), exhibited no significant difference between the groups (data not shown).
ST3GAL5 is ubiquitously distributed throughout the body and is particularly abundant in the central nervous system.1920 To date, a few researchers have investigated the association between genetic variants of ST3GAL5 and human disease. Animal and clinical studies have shown that loss or decreased function of ST3GAL5 is associated with enhanced insulin sensitivity, early-onset epilepsy, and complete hearing loss.21222324 ST8SIA1 is mainly expressed in the brain, kidney, thymus, and testes.19 Previous study reported that several SNPs of ST8SIA1 were significantly associated with susceptibility to multiple sclerosis, while another study reported that there was no significant association between multiple sclerosis and these SNPs in their study populations.2526
Here, we investigated the functional effect of genetic variants in ST3GAL5 and ST8SIA1 using in vitro molecular assays. Three ST3GAL5 promoter haplotypes, H1, H3, and H4, exhibited a significant decrease in promoter activity: H3 had the most significant decrease (45%). Decreased activities of these haplotypes were due to two genetic variants, g.-1468C>G and g.-1051G>C, present in H1, H3, and H4. Previous studies have reported that a number of transcription factors, AP-4, myeloid zinc finger-1, SP1, and cAMP-responsive element binding protein (CREB), regulate the transcription of ST3GAL5.272829 Among them, Chung, et al.29 reported that CREB is activated by protein kinase C and the extracellular regulated kinase signal transduction pathway, and induces ST3GAL5 expression. Th-rough TFBS analysis and EMSA, we found that a transcription factor, AP-1, regulates ST3GAL5 transcription. AP-1 is a strong oncogene and is expressed in a variety of tumors.30 Previous studies have reported that AP-1 acts bifunctionally as an inducer or repressor for the transcription of various genes.1631323334 In this study, we observed that the ST3GAL5 promoter contains the sequence, CGTGAGA, similar to a/gc/gTGACT, the consensus sequence recognized by AP-1. The variant g.-1051G>C results in the sequence, CGTGACA, which more closely resembles the AP-1 consensus sequence. Based on this observation, we predicted that AP-1 would preferentially bind to the variant DNA sequence over the reference; this prediction was indeed confirmed by EMSA. We further examined the effect of AP-1 on the ST3GAL5 promoter activity by using siRNA to knockdown c-Jun, one of components of AP-1. As a result, we observed that a significant increase in promoter activity of the reference (H2) and g.-1051G>C reporter constructs in the presence of c-Jun siRNA, compared with the promoter activities with the negative control siRNA, by 50.55 and 76.66%, respectively (Supplementary Fig. 2, only online). This result suggests that AP-1 works as a repressor of ST3GAL5 transcription. Regarding ST8SIA1, Kang, et al.35 reported that the ST8SIA1 promoter contains binding sites for transcriptional factors, including C-Ets-1, CREB, AP-1, and NF-κB, which play an important role in the induction of ST8SIA1 expression in Fas-activated Jurkat T cells. In the present study, H2, a haplotype of ST8SIA1, showed a significant increase in promoter activity, and two transcriptional factors, NKX3.1 and SP1, were identified as playing a role in the regulation of ST8SIA1 activity. NKX3.1 is known as a tumor suppressor gene of prostatic cancer, and a functional variant in this gene is associated with increased prostate cancer risk.3637 SP1 is ubiquitously expressed and works as a housekeeping transcription factor.38 For example, it plays a role in the insulin regulation of gene expression.39 Previous study reported that the promoter of mouse lysosomal sialidase, which plays a role in the catabolism of gangliosides, is responsive to SP1.40
Previously, we observed that mRNA levels of ST3GAL5 and ST8SIA1 increased in orbital fat tissues obtained from TAO patients, compared with healthy controls.11 In the present study, we could not find TAO-specific ST3GAL5 or ST8SIA1 variants: all identified genetic variants were found in the healthy subjects. Multifactorial mechanisms have been suggested to be involved in the pathogenesis of TAO.4 Taken together with the clinical phenomenon that not all GD patients present with TAO, future studies including patients with GD who have no sign of TAO are needed to be performed. In terms of the severity of this disease, we observed that patients with the ST8SIA1 haplotype H2, which showed an increase of promoter activity, showed less severe exophthalmos, compared with patients without H2. Moreover, although it was not statistically significant, TAO patients without ST3GAL5 haplotypes H1, H3, and H4 that showed an increase of promoter activity also showed a tendency to experience less severe exophthalmos, compared with the other group. In patients with TAO, proptosis develops as a result of the increased volume of orbital tissues, such as extraocular muscle and orbital fat, which is caused by the edematous change. Deposition of HA is one of the pathologic changes in the orbital tissue of patients with TAO and is believed to be one of the causes of tissue edema from the hydrophilic characteristic of HA. Taken together with previous findings showing that GT1b increase HA11 and that HA induces cyclooxygenase-2 in orbital fibroblasts,41 the present results contradict the expectation that patients possessing haplotypes of ST3GAL5 or ST8SIA1 with increased promoter activity would show more severe clinical phenotypes. One possible explanation for this discrepancy might be the involvement of other mechanisms that modulate the synthesis and/or secretion of GT1b and/or HA in orbital fibroblasts, as illustrated by the divergent effects of IL-4 and interferon-γ on IL-1β-induced HA and prostaglandin E2 production.42 In addition, in vivo, orbital fibroblasts in TAO receive signals from a combination of various mediators, and possibly interfere with each other's individual biological effects, so that the net biological effect of orbital fibroblast activating factors in the pathophysiology of TAO is complex.
Limitations of this study include the insufficient sample size used in the analysis of the clinical data to achieve strong statistical power, and the lack of actual observations of promoter activity depending on the presence of genetic variants in the orbital tissues of the corresponding patients. Therefore, to validate the relevance of the functional characteristics of the identified genetic variants to the clinical characteristics of TAO patients, more association studies with a large number of samples, including the orbital tissues, should be performed. Another limitation is that we could not examine the effect of the differences in the promoter activities of ST3GAL5 or ST8SIA1 depending on its variants on the actual expression level and/or activity of these enzymes. To the best of our knowledge, no study has been reported in which the activity of ST3GAL5 or ST8SIA1 was measured. Alternatively, the correlation between the transcriptional activity and expression levels of ST3GAL5 or ST8SIA1 has been suggested in a couple of studies.2943 In human breast cancer cells, estradiol decreases ST8SIA1 mRNA expression by repression of its promoter activity through prevention of NF-κB binding to the ST8SIA1 promoter.43 In our current study, the haplotypes of ST3GAL5 or ST8SIA1 markedly changed the luciferase activity of each gene (13.45–102.30%). Taken together with our results showing the mechanism underlying the transcriptional regulation of the ST3GAL5 or ST8SIA1 promoter, these findings suggest that the change in promoter activity exhibited by ST3GAL5 or ST8SIA1 variants might ultimately affect the expression level and/or activity of ST3GAL5 or ST8SIA1.
In conclusion, we identified and characterized genetic variants in the promoter regions of ST3GAL5 and ST8SIA1 in Korean individuals. Our genotype-phenotype analysis data might suggest a possible link between the ST8SIA1 functional promoter haplotype and the severity of TAO. To the best of our knowledge, this is the first study to have examined the effect of ST3GAL5 and ST8SIA1 promoter variants on the transcription of genes and the relationship between haplotypes and clinical phenotypes of TAO in Koreans. Further studies are warranted to investigate whether these functional haplotypes of ST3GAL5 and ST8SIA1 promoters are associated with clinical features of other diseases related to ganglioside synthesis.
Figures and Tables
 | Fig. 1Effect of genetic variants on the promoter activity of ST3GAL5 or ST8SIA1. Luciferase activities were measured 30 hours after transfection of the reporter vectors containing ST3GAL5 (A and B) or ST8SIA1 (C and D) haplotypes (A and C) or variants (B and D) into HCT-116 cells. The luciferase activity of each construct was compared with that of the reference (REF). The data (mean±SD) were obtained from triplicate wells. *p<0.01, †p<0.001. |
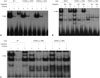 | Fig. 2Effect of genetic variants on the binding of the transcription factors to ST3GAL5 (A) or ST8SIA1 (B and C) promoters. (A) 32P-labelled oligonucleotides (lanes 1-3, AP-1 consensus; lanes 4-6, g.-1051G reference; lanes 7-9, g.-1051C variant) were incubated with nuclear protein extracts (25 µg) obtained from HCT-116 cells. Competition assay was performed using 100-fold molar excess of unlabeled AP-1 consensus oligonucleotides (lanes 2, 5, and 8). Supershift assays were performed with an antibody against phosphorylated c-Jun (lanes 3, 6, and 9). (B) 32P-labelled oligonucleotides (lanes 1-3, NKX3.1 consensus; lanes 4-6, g.-2196A reference; lanes 7-9, g.-2196C variant) were incubated with nuclear protein extracts (25 µg) obtained from HCT-116 cells. Competition assay was performed using 100-fold molar excess of unlabeled NKX3.1 consensus (Ref, lanes 2, 5, and 8) or core sequence-mutated (Mut, lanes 3, 6, and 9) oligonucleotides. (C) 32P-labelled oligonucleotides (lanes 1-3, SP1 consensus; lanes 4-6, g.-1984T reference; lanes 7-9, g.-1984C variant) were incubated with nuclear protein extracts (25 µg) obtained from HCT-116 cells. Competition assay was performed using 100-fold molar excess of unlabeled SP1 consensus oligonucleotides (lanes 2, 5, and 8). Supershift assays were performed with an antibody against SP1 (lanes 3, 6, and 9). The arrows indicate the position of the DNA-protein complex. |
 | Fig. 3Effect of a nonsynonymous variant, H104R, on ST3GAL5 expression. Immunoblotting assays were performed using cell lysates obtained 48 hours after the transfection of empty vector (EV, pcDNA3.1), ST3GAL5 reference (REF), or variant plasmids into HCT-116 cells. The data (mean±SD) were obtained from three representative experiments. β-actin was used as an internal control. |
Table 1
Components of the Clinical Activity Score
| Spontaneous retrobulbar pain |
| Pain on attempted upward or downward gaze |
| Redness of eyelids |
| Redness of conjunctiva |
| Swelling of caruncle or plica |
| Swelling of eyelids |
| Swelling of conjunctiva (chemosis) |
Table 2
Frequency of ST3GAL5 or ST8SIA1 Genetic Variations in Thyroid-Associated Ophthalmopathy Patients

Table 3
Frequency of ST3GAL5 Promoter Haplotypes

| ID | g.-1930A>G | g.-1626G>A | g.-1468C>G | g.-1051G>C | g.-141T>C | g.-140C>A | Frequency |
|---|---|---|---|---|---|---|---|
| H1 | G | G | C | C | C | A | 0.510 |
| H2 | A | G | C | G | T | C | 0.260 |
| H3 | A | A | G | C | C | C | 0.130 |
| H4 | A | G | C | C | C | C | 0.100 |
Table 4
Frequency of ST8SIA1 Promoter Haplotypes

| ID | g.-2196A>C | g.-1984T>C | g.-681G>C | Frequency |
|---|---|---|---|---|
| H1 | A | T | G | 0.900 |
| H2 | C | C | G | 0.100 |
Table 5
Comparison of Clinical Characteristics According to ST3GAL5 Haplotypes

Table 6
Comparison of Clinical Characteristics According to the ST8SIA1 Haplotype

ACKNOWLEDGEMENTS
We thank all the participants in this study. This study was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (2010-0027945) (JH Choi), (NRF-2016R1A2B1014704) (KH Kook) and the Ministry of Education (NRF-2012R1A1A1038670) (JH Choi).
Notes
References
1. Kuriyan AE, Phipps RP, Feldon SE. The eye and thyroid disease. Curr Opin Ophthalmol. 2008; 19:499–506.

2. RUNDLE FF. Management of exophthalmos and related ocular changes in Graves' disease. Metabolism. 1957; 6:36–48.
3. Wiersinga WM, Bartalena L. Epidemiology and prevention of Graves' ophthalmopathy. Thyroid. 2002; 12:855–860.

5. Bartalena L, Pinchera A, Marcocci C. Management of Graves' ophthalmopathy: reality and perspectives. Endocr Rev. 2000; 21:168–199.

6. Liu YH, Chen CC, Liao LL, Wan L, Tsai CH, Tsai FJ. Association of IL12B polymorphisms with susceptibility to Graves ophthalmopathy in a Taiwan Chinese population. J Biomed Sci. 2012; 19:97.

7. Liu YH, Chen RH, Wu HH, Liao WL, Chen WC, Tsai Y, et al. Association of interleukin-1β (IL1B) polymorphisms with Graves' ophthalmopathy in Taiwan Chinese patients. Invest Ophthalmol Vis Sci. 2010; 51:6238–6246.

8. Niyazoglu M, Baykara O, Koc A, Aydogğdu P, Onaran I, Dellal FD, et al. Association of PARP-1, NF-κB, NF-κBIA and IL-6, IL-1β and TNF-α with Graves disease and Graves ophthalmopathy. Gene. 2014; 547:226–232.

9. Liao WL, Chen RH, Lin HJ, Liu YH, Chen WC, Tsai Y, et al. Toll-like receptor gene polymorphisms are associated with susceptibility to Graves' ophthalmopathy in Taiwan males. BMC Med Genet. 2010; 11:154.

10. van Echten G, Sandhoff K. Ganglioside metabolism. Enzymology, topology, and regulation. J Biol Chem. 1993; 268:5341–5344.

11. Kook KH, Choi YH, Kim YR, Park SJ, Jou I, Kim SJ, et al. Altered ganglioside expression modulates the pathogenic mechanism of thyroid-associated ophthalmopathy by increase in hyaluronic acid. Invest Ophthalmol Vis Sci. 2011; 52:264–273.

12. Kaback LA, Smith TJ. Expression of hyaluronan synthase messenger ribonucleic acids and their induction by interleukin-1β in human orbital fibroblasts: potential insight into the molecular pathogenesis of thyroid-associated ophthalmopathy. J Clin Endocrinol Metab. 1999; 84:4079–4084.

13. Werner SC. Classification of the eye changes of Grave's disease. J Clin Endocrinol Metab. 1969; 29:982–984.

14. Mourits MP, Prummel MF, Wiersinga WM, Koornneef L. Clinical activity score as a guide in the management of patients with Graves' ophthalmopathy. Clin Endocrinol (Oxf). 1997; 47:9–14.

15. Lam AK, Lam CF, Leung WK, Hung PK. Intra-observer and inter-observer variation of Hertel exophthalmometry. Ophthalmic Physiol Opt. 2009; 29:472–476.

16. Choi JH, Yee SW, Kim MJ, Nguyen L, Lee JH, Kang JO, et al. Identification and characterization of novel polymorphisms in the basal promoter of the human transporter, MATE1. Pharmacogenet Genomics. 2009; 19:770–780.

17. Choi JH, Yee SW, Ramirez AH, Morrissey KM, Jang GH, Joski PJ, et al. A common 5'-UTR variant in MATE2-K is associated with poor response to metformin. Clin Pharmacol Ther. 2011; 90:674–684.

18. Jang GH, Kim TH, Choe Y, Ham A, Choi JH. Functional characterization of genetic variations in the MDR3 promoter. Biochem Biophys Res Commun. 2013; 430:1312–1318.

19. Tsuji S. Molecular cloning and functional analysis of sialyltransferases. J Biochem. 1996; 120:1–13.

21. Yamashita T, Hashiramoto A, Haluzik M, Mizukami H, Beck S, Norton A, et al. Enhanced insulin sensitivity in mice lacking ganglioside GM3. Proc Natl Acad Sci U S A. 2003; 100:3445–3449.

22. Yoshikawa M, Go S, Takasaki K, Kakazu Y, Ohashi M, Nagafuku M, et al. Mice lacking ganglioside GM3 synthase exhibit complete hearing loss due to selective degeneration of the organ of Corti. Proc Natl Acad Sci U S A. 2009; 106:9483–9488.

23. Simpson MA, Cross H, Proukakis C, Priestman DA, Neville DC, Reinkensmeier G, et al. Infantile-onset symptomatic epilepsy syndrome caused by a homozygous loss-of-function mutation of GM3 synthase. Nat Genet. 2004; 36:1225–1229.

24. Fragaki K, Ait-El-Mkadem S, Chaussenot A, Gire C, Mengual R, Bonesso L, et al. Refractory epilepsy and mitochondrial dysfunction due to GM3 synthase deficiency. Eur J Hum Genet. 2013; 21:528–534.

25. Husain S, Yildirim-Toruner C, Rubio JP, Field J, Schwalb M, et al. Southern MS Genetics Consortium. Variants of ST8SIA1 are associated with risk of developing multiple sclerosis. PLoS One. 2008; 3:e2653.

26. Ramagopalan SV, Morrison KM, Para A, Handel A, Disanto G, Handunnetthi L, et al. Variants in ST8SIA1 do not play a major role in susceptibility to multiple sclerosis in Canadian families. J Neuroimmunol. 2009; 212:142–144.

27. Kim SW, Lee SH, Kim KS, Kim CH, Choo YK, Lee YC. Isolation and characterization of the promoter region of the human GM3 synthase gene. Biochim Biophys Acta. 2002; 1578:84–89.

28. Zeng G, Gao L, Xia T, Tencomnao T, Yu RK. Characterization of the 5'-flanking fragment of the human GM3-synthase gene. Biochim Biophys Acta. 2003; 1625:30–35.

29. Chung TW, Choi HJ, Lee YC, Kim CH. Molecular mechanism for transcriptional activation of ganglioside GM3 synthase and its function in differentiation of HL-60 cells. Glycobiology. 2005; 15:233–244.

30. Ozanne BW, Spence HJ, McGarry LC, Hennigan RF. Transcription factors control invasion: AP-1 the first among equals. Oncogene. 2007; 26:1–10.

31. Pfäfflin A, Brodbeck K, Heilig CW, Häring HU, Schleicher ED, Weigert C. Increased glucose uptake and metabolism in mesangial cells overexpressing glucose transporter 1 increases interleukin-6 and vascular endothelial growth factor production: role of AP-1 and HIF-1α. Cell Physiol Biochem. 2006; 18:199–210.

32. Duane WC, Xiong W, Lofgren J. Transactivation of the human apical sodium-dependent bile acid transporter gene by human serum. J Steroid Biochem Mol Biol. 2008; 108:137–148.

33. Ramírez-Sotelo G, López-Bayghen E, Hernández-Kelly LC, Arias-Montaño JA, Bernabé A, Ortega A. Regulation of the mouse Na+-dependent glutamate/aspartate transporter GLAST: putative role of an AP-1 DNA binding site. Neurochem Res. 2007; 32:73–80.

34. Mittelstadt ML, Patel RC. AP-1 mediated transcriptional repression of matrix metalloproteinase-9 by recruitment of histone deacetylase 1 in response to interferon β. PLoS One. 2012; 7:e42152.

35. Kang NY, Kang SK, Lee YC, Choi HJ, Lee YS, Cho SY, et al. Transcriptional regulation of the human GD3 synthase gene expression in Fas-induced Jurkat T cells: a critical role of transcription factor NF-κB in regulated expression. Glycobiology. 2006; 16:375–389.

36. Martinez EE, Darke AK, Tangen CM, Goodman PJ, Fowke JH, Klein EA, et al. A functional variant in NKX31 associated with prostate cancer risk in the Selenium and Vitamin E Cancer Prevention Trial (SELECT). Cancer Prev Res (Phila). 2014; 7:950–957.

37. Song LN, Silva J, Koller A, Rosenthal A, Chen EI, Gelmann EP. The tumor suppressor NKX3.1 is targeted for degradation by DYRK1B kinase. Mol Cancer Res. 2015; 13:913–922.

38. Solomon SS, Majumdar G, Martinez-Hernandez A, Raghow R. A critical role of Sp1 transcription factor in regulating gene expression in response to insulin and other hormones. Life Sci. 2008; 83:305–312.

39. Samson SL, Wong NC. Role of Sp1 in insulin regulation of gene expression. J Mol Endocrinol. 2002; 29:265–279.

40. Champigny MJ, Johnson M, Igdoura SA. Characterization of the mouse lysosomal sialidase promoter. Gene. 2003; 319:177–187.

41. Lim HS, Back KO, Kim HJ, Choi YH, Park YM, Kook KH. Hyaluronic acid induces COX-2 expression via CD44 in orbital fibroblasts from patients with thyroid-associated ophthalmopathy. Invest Ophthalmol Vis Sci. 2014; 55:7441–7450.

42. Han R, Smith TJ. T helper type 1 and type 2 cytokines exert divergent influence on the induction of prostaglandin E2 and hyaluronan synthesis by interleukin-1β in orbital fibroblasts: implications for the pathogenesis of thyroid-associated ophthalmopathy. Endocrinology. 2006; 147:13–19.

43. Bobowski M, Vincent A, Steenackers A, Colomb F, Van Seuningen I, Julien S, et al. Estradiol represses the GD3 synthase gene ST8SIA1 expression in human breast cancer cells by preventing NFκB binding to ST8SIA1 promoter. PLoS One. 2013; 8:e62559.
SUPPLEMENTARY MATERIALS
Supplementary Fig. 1
Electrophoretic mobility shift assay for v-Myb in, ST3GAL5 g-1468C>G variant. 32P-labelled oligonucleotides (lanes 1–3, v-Myb consensus; lanes 4–6, g.-1468C reference; lanes 7–9, g.-1468G variant) were incubated with nuclear protein extracts (25 µg) obtained from HCT-116 cells. Competition assay was performed using 50- (lanes 2, 5, and 8) or 100-fold (lanes 3, 6, and 9) molar excess of unlabeled v-Myb consensus oligonucleotides. The arrow indicates the position of the DNA-protein complex.
Supplementary Fig. 2
Effect of AP-1 on the ST3GAL5 transcription. After 24 hours of siRNA transfection to knockdown AP-1, the reference (g.-1051G) or variant (g.-1051C) reporter constructs were transfected into HCT-116 cells. Then, luciferase activities were determined after 48 hours. **p<0.01 vs. the luciferase activity with the negative control siRNA, †p<0.05, ‡p<0.01 vs. the luciferase activity without the siRNA. EV, empty vector; siRNA, small interfering RNA.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download