Abstract
Purpose
The genes for cytochrome P450 2C9 (CYP2C9) and vitamin K epoxide reductase complex subunit 1 (VKORC1) have been identified as important genetic determinants of warfarin dosing and have been studied. We developed warfarin algorithm for Korean patients with stroke and compared the accuracy of warfarin dose prediction algorithms based on the pharmacogenetics.
Materials and Methods
A total of 101 patients on stable maintenance dose of warfarin were enrolled. Warfarin dosing algorithm was developed using multiple linear regression analysis. The performance of all the algorithms was characterized with coefficient of determination, determined by linear regression, and the mean of percent deviation was used to predict doses from the actual dose. In addition, we compared the performance of the algorithms using percentage of predicted dose falling within ±20% of clinically observed doses and dividing the patients into a low-dose group (≤3 mg/day), an intermediate-dose group (3–7 mg/day), and high-dose group (≥7 mg/day).
Warfarin, the most commonly used oral anticoagulant in the world, is indicated for the prevention and treatment of thromboembolic events in patients with deep vein thrombosis, pulmonary embolism, atrial fibrillation and prosthetic heart valves. It is administered as a racemic mixture of S-warfarin and R-warfarin. S-warfarin is the more active isomer and has a greater therapeutic effect. These enantiomers are extensively metabolized by various cytochrome P450 enzymes. R-warfarin is mainly metabolized by cytochrome P450 1A2 (CYP1A2), CYP2C19, and CYP3A4, while S-warfarin is predominantly metabolized by CYP2C9.1 Both enantiomers affect the coagulation cascade by inhibiting the activity of vitamin K epoxide reductase complex 1 (VKORC1), thus interfering with the activation of clotting factors II, VII, IX, and X.2 However, despite its considerable benefit, warfarin is less frequently prescribed than it should be,3 because of it's relatively high adverse event rate and the difficulty in managing the therapy.4 Common adverse drug events (ADEs) arising from warfarin treatment include major and minor bleeding and hemorrhagic stroke.5 ADEs are also frequently due to insufficient therapy: ischemic stroke and venous thromboembolism.6 Warfarin, which has narrow therapeutic index, shows not only large inter-individual variations in dose response but also intra-individual variation. Because patients' clinical characteristics such as age, gender, body weight, concurrent medications, diet, co-morbidities and patient compliance level have shown to have large influence on warfarin dosing,7 frequent monitoring of its effect, as measured by the international normalized ratio (INR), is warranted.
CYP2C9 and VKORC1 have been identified as important genetic determinants of warfarin dosing. The most common CYP2C9 genotype among all ethnic is CYP2C9*1 and found in about 80% of Caucasians8 and 93% of Korean.9 Lindh, et al.10 demonstrated that carriers of CYP2C9*2 and CYP2C9*3 alleles require less warfarin dose than carriers of wild type CYP2C9*1 genotype. Difference in allelic frequencies are also observed with the most common single nucleotide polymorphisms (SNPs) in the VKORC1 gene, 1173C>T (rs9934438). Approximately, 35% of caucasians carry CC genotype, while only about 15% carry the TT genotype. However, most of the Korean showed TT genotype, but less than 1% carry CC genotype.11 Carriers of 1173CC and 1173CT genotype need 44% and 97%, respectively, more warfarin dose than carriers of 1173TT genotype.12 The clinical algorithms for determining warfarin dose containing clinical characteristics and pharmacogenetics information of the patients have been developed for more proper warfarin dose prediction. These algorithms are not intended to replace INR monitoring, but to increase the accuracy and reduce trial and error approach in warfarin dosing. According to International Warfarin Pharmacogenetics Consortium (IWPC) study,13 the algorithms using clinical and genetic information increase the accuracy in dose prediction than a fixed-dose approach or algorithms derived from clinical information. In addition, they are useful, especially in the patients who may be administered more or less than appropriate dose.13
Personalized dosing and INR monitoring are required, because response to warfarin is different depending on the indication and the state of the disease. The studies on warfarin dose assessment so far was mainly targeted at valvular heart disease, deep vein thrombosis and atrial fibrillation.131415161718192021 On the other hand, warfarin is widely used to prevent the recurrence of stroke, which occupied the second place in the current causes of death in Korea, nevertheless, there are only a few studies to compare the predictive power of the dosing control based on a pharmacogenetics. This study reviewed prescribed dose and actual INR response in patients with stroke and compared the accuracy of 10 warfarin dose prediction algorithms based on the pharmacogenetics. In addition, warfarin dosing algorithm for Korean patients with stroke was developed to increase the quality of care for stroke patients.
A total of 129 patients undergoing warfarin treatment for prevention and treatment of stroke and requesting genotyping of CYP2C9 and VKORC1 were recruited retrospectively at the neurology clinic at the Severance Hospital, Seoul, Korea. All study participants were enrolled between January 2009 and December 2014. Patients included were adults, whose warfarin dose requirement had remained constant for at least 3 previous clinic visits over a minimum period of 3 months, and with an INR of the prothrombin time within the range of 1.5 to 3.0.22 Twenty-eight patients were excluded from the study according to enrolment criteria. This study was approved by the Institutional Review Board of the Yonsei University Severance Hospital, Seoul, Korea. Written informed consent was obtained from the patients.
Data were collected from patients' medical records. These data included demographic characteristics, comorbidities, the stable therapeutic dose of warfarin, the INR achieved with a stable warfarin dose, the use of concomitant medications, and the genotype of CYP2C9 and VKORC1. The interacting drugs, which is defined based on previously published literature, were also reviewed.2324
Genomic DNA was extracted from EDTA whole blood samples with QIAamp DNA Extraction Kit (Qiagen, Hilden, Germany). For determination of the CYP2C9 genotype, the CYP2C9*3 (1075A>C; rs1057910) SNP is selected. For VKORC1 genotypes, the VKORC1 1173C>T (rs 9934438) SNP is determined. PCR and direct sequencing were performed using primers designed in Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/). Purified PCR products were obtained using a QIAquick Gel Extraction Kit (Qiagen, Düsseldorf, Germany) and were sequenced using a Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA). Sequences were analyzed using an ABI 3500dx system (Applied Biosystems). To detect any sequence variation, the sequences were compared to the reference sequences using Sequencher software (Gene Codes, Ann Arbor, MI, USA).
A literature search from Pubmed database was performed, with the search terms warfarin, algorithm, polymorphism, CYP2C9 and VKORC1, to select warfarin dosing algorithms. Algorithms were included based on the following criteria; 1) equations to predict maintenance warfarin dose, 2) only two SNPs consisting of VKORC1 1173G>T (or VKORC1 -1639G>A and 2255C>T) and CYP2C9*2 and/or *3, 3) published in English, 4) equations containing available clinical parameters, 5) algorithms were selected, depending on a historical aspect (e.g., the first warfarin dosing model), the size or ethnicity of the study population. Studies that enrolled adult patients with atrial fibrillation, venous thromboembolic diseases, recent orthopedic surgery, valvular disease, and stroke were also included. Wherever genotype is missing, we imputed its value based on other VKORC1 SNPs, because VKORC1 1173 genotype is suggested to be in complete linkage disequilibrium with VKORC1 -1639 and 2255.1325
Warfarin doses between the different genotypes were compared using the Mann Whitney U-test. Predictive accuracy was assessed by comparing the dose predicted by the nine algorithms to actual dose which the patient was taking. Predicted dose was calculated using published equations, except Gage's calculated by input on the website http://www.warfarindosing.org. A best fit trendline and correlation coefficient were determined by linear regression. In addition, the mean of percent deviation of predicted doses from the actual dose was used to evaluate the predictive accuracy of each algorithms. In addition, we compared the performance of the algorithms using percentage of predicted dose falling within ±20% of clinically observed doses26 and dividing the patients into low-dose group (≤3 mg/day), intermediate-dose group (3–7 mg/day), and high-dose group (≥7 mg/day).13 Stepwise multiple regression analysis was performed to develop new warfarin dosing algorithm, and the results of univariate analysis were used to choose predictors for multivariate analysis. All statistical tests were performed with a p value<0.05 significance. All analyses were performed using the Statistical Package for Social Science (SPSS 18.0; SPSS Inc., Chicago, IL, USA).
The characteristics of all 101 patients are listed in Table 1. A total of 101 patients with a mean age 64 years (SD, ±13.4) ranging from 27 to 88 years, included 64 males (63%). The mean body weight was 64.6 kg (SD, ±10.5), and the mean body surface area was 1.7 (SD, ±0.2). The mean stable warfarin dose was 3.74 mg/day (SD, ±1.43). Concurrent diseases associated with these patients included atrial fibrillation (63 patients, 62%), hypertension (52 patients, 52%), diabetes mellitus (27 patients, 27%), heart disease including coronary arterial occlusive disease (14 patients, 14%), heart failure (7 patients, 7%), and cardiac valvular disease (9 patients, 9%). Other sources of cardioemboilism including patent foramen ovale and left atrial thrombus were identified, in addition to heart problem that is shown in the Table 1. A total of 44 (44%) patients were receiving comedication that could affect the anticoagulation effect of warfarin, including amiodarone, aspirin, antiplatelet drugs, statins, thyroid hormone, and verapamil.
Table 2 shows daily warfarin dose of different genotypes. For CYP2C9, 97 patients (96%) were identified to be homozygous for CYP2C9*1, and 4 patients (4%) were heterozygous for CYP2C9*3. No patients with CYP2C9*2 allele were identified. The frequency of the VKORC1 1173TT genotype was 82% and that of 1173CT genotype was 17%, and only 1 patient was homozygous for the variant C allele. In our study, no patients with homozygous CYP2C9*3/*3 were identified, and the CYP2C9 and VKORC SNPs for Korean were in Hardy-Weinberg equilibrium. The stable warfarin doses for patients with VKORC1 TT type (3.6±1.2 mg/day) were significantly lower than those for patients with any other VKORC1 genotype (p<0.05). However, the difference in the stable warfarin dose between patients with homozygous for CYP2C9*1 (3.8±1.4 mg/day) and heterozygous for CYP2C9*3 (2.6±0.5 mg/day) was not significant.
For multiple linear regression analysis, 4 variables, including age, bodyweight, CYP2C9*3 and VKORC1 1173 genotypes, were selected (R2=0.51) (Table 3). We established the warfarin dosing formula with following equation: maintenance dose=exp [1.756-0.015 (age)+0.006 (body weight)-0.284 (CYP2C9 genotype)+ 0.407 (VKORC1 genotype)]. It was coded as 1 in the case of the presence of the CYP2C9 variant, or the presence of the VKORC1 1173C allele.
A comparison of the ten algorithms for determining warfarin doses is shown in Table 4. Most algorithms that evaluated, including the dosing algorithm derived from this study, had a good correlation. However, the algorithms by Gage, et al.,14 Wu, et al.,21 and Huang, et al.19 showed poor correlation. Among these 7 algorithms, algorithms from this study, Sconce, et al.,15 Anderson, et al.,20 and Ohno, et al.16 produce similar accuracy with mean deviation ranging from -10.0 to 4.3. These algorithms were selected based on their correlation coefficient (r>0.6) and the mean deviation from the actual dose (mean deviation about 10%) for further analysis. The algorithm from this study provided significantly better prediction fell within 20% of the actual dose (Table 5). Others performed similarly to predict ideal dose. Algorithms by Sconce, et al.15 and Ohno, et al.16 tend to underestimate in about 40% of cases. In addition, the accuracy of this study and Anderson, et al.20 was better than others for patients who need intermediate-dose group (Table 6). For patients who need less than 3 mg/day, algorithm by Ohno performed well. However, for patients who required more than 7 mg/day, all algorithms performed poorly, with underestimation for all patients.
Warfarin, the first human anticoagulant, remains the most commonly prescribed oral anticoagulant in the world. Warfarin exerts its effect by inhibiting the activity of VKORC1, thus interfering with the activation of vitamin K-dependent clotting factors II, VII, IX, and X.2 Warfarin is underutilized for stroke prevention. The Agency for Healthcare Policy and Research noted that physicians avoid to prescribe warfarin, because they are not familiar with techniques for administrating the drug safely and fear bleeding complication.34 Warfarin therapy is challenging, because warfarin has narrow therapeutic index. In addition, it shows not only large inter-individual variations in dose response but also intra-individual variation. Because patients' clinical characteristics such as age, gender, body weight, concurrent medications, diet, co-morbidities and patient compliance level largely influence warfarin dosing,7 frequent monitoring of its effect, as measured by the INR, is warranted.
CYP2C9 and VKORC1 have been identified as important genetic determinants of warfarin dosing. Two prospective studies2027 on genotype-guided warfarin dosing predicted more accurately, resulting in reduction of dosing changes, minor bleeding complication, and time to reach therapeutic range. Although numerous warfarin dosing algorithms have been developed, their indications for warfarin usage were heterogeneous. Until now, there has been no consensus pharmacogenetic-guided algorithm.
In the present study, we developed an algorithm to provide a practical warfarin dosing for Korean patients with stroke. The warfarin dosing algorithm was developed on a homogeneous population and single disease indication for stroke, since warfarin have been underused for prevention of stroke and there are few studies on warfarin dosing algorithm focused on stroke patients. Because the distribution of warfarin dose is skewed, we created dosing algorithm for log transformation of doses, as evidenced by a mean percent deviation that was lower than that for both the raw doses square root of doses. Other variables including age and body weight showed normal distribution. We analyzed whether ten selected dosing algorithms, including the algorithm derived in the present study, could accurately predict warfarin dose in the study population, and found that the present algorithm demonstrated good correlation with actual dose, with coefficient of determination (R2) of 0.51. Algorithm in this study is consisted of five factors; age, body weight, and genotypes of CYP2C9 and VKORC1. While reduced incorporated factors are convenient for physicians to use, this algorithm performed better than Gage, et al.,14 Wu, et al.,21 and Huang, et al.19 The VKORC1 and CYP2C9 genotypes accounted for about 14% of the inter-individual variation of the maintenance daily warfarin dose. Because allele frequencies of VKORC1 and CYP2C9 are different depending on race, the R2 values of these genes differ among studies.
Anderson, et al.,20 Ohno, et al.,16 and Sconce, et al.15 also showed good linear relationship with actual dose and predicted dose. However, the R2 indicated the linearity of the association only, and the mean deviation from actual dose is a better measure of the algorithms' performance. Although the above three algorithms showed a good correlation with the actual dose in our study population, a better prediction of dosage was achieved by our model.
The algorithm devised by Cho, et al.18 was the latest warfarin dosing algorithm for Korean patients with atrial fibrillation and the best model for prediction of daily maintenance dose. This algorithm showed a good relationship between the actual dose and the predicted warfarin dose in our study population as reported previously. However, this algorithm was the worst performing algorithm by means of the mean deviation. The mean age, the strongest predictor of warfarin dose, was slightly older in cohort of Cho, et al., although its significance is unclear. Because these two algorithms developed for two different single disease indication; atrial fibrillation and stroke, these patients may be differently influenced by environmental factors such as coadministered drugs and comorbidities. Atrial fibrillation was indeed the most common indication of warfarin usage in this study. Likewise western countries,28 cervicocephalic artery dissections were common causes of stroke in young patients under 45. Cho, et al.18 reported that statin influences the daily dose of warfarin. Simvastatin, fluvastatin, and lovastatin potentiate warfarin's effect.2429 In our study, most patients were taking statin such as atorvastatin, pitavastatin, and rosuvastatin which do not affect the warfarin's effect. There is no correlation between daily dose of warfarin and statin status regardless of types of statin.
The algorithm derived in the present this study was less predictable among patients who required high doses of warfarin (≥7 mg/day). As this study was a small retrospective analysis with only a few patients requiring high doses, the results might have been skewed because of the individual patients variations. About 3% of the patients could have complication due to underdose. Of the three outliers, two patients were VKORC1 CT genotype. The other was TT type and he was taking antituberculosis drug. Rimfampin decreases INR increase via induction of hepatic metabolism of warfarin.24 Removal of these three data points did not improve the correlation coefficient for our algorithm (R2=0.51). However, the sample size was too small to make a conclusion on the efficacy of the dosing algorithms in this population.
In this study, we found 0 and 4% prevalence of CYP2C9*2 and CYP2C9*3, respectively, which is consistent with the report of Cho, et al. who also found no CYP2C9*2 and a 8.5% prevalence of *3. For VKORC1, we found 82%, 17%, and 1% prevalence of VKORC1 TT, CT, CC genotype, respectively, which compares with the report of Cho, et al. who found 75.4%, 23.1%, and 1.5%, respectively.
Our study has several limitations. First, we did not have sufficient data to include potentially important factors such as vitamin K intake or compliance of administration even if we educated the patients when starting warfarin. However, the percentage of variability in warfarin dosing in our study is similar to that in other compared models, therefore, the effect of these variables is probably small. Second, because we investigated only one VKORC1 SNPs, requiring us to impute missing genotype for evaluating some models. Therefore, we substituted missing genotype based on linkage disequilibrium, which is generally reliable.25 Nevertheless, it may cause error that would lead to decrease of the accuracy of our model. Third, only 4% of the study population was younger than 40 years of age; therefore, additional models for stroke patients with younger age are needed, as age is important factor of prediction.
In order to further improve dosing algorithms, additional study is necessary in efforts to find new genes and SNPs contained with these genes that influence warfarin pharmacokinetics and pharmacodynamics. Although the incorporation of additional variables could improve predictive algorithm, the gains may be modest and probably do not justify the cost effectiveness and improvement of clinical outcome. In addition, studies on clinical utility of these pharmacogenetic-guided algorithms should be performed.
In conclusion, we developed warfarin dose prediction algorithm for patients with stroke, and it explained 51% of the variation in the daily maintenance warfarin dose. Further studies to elucidate clinical utility of genotype-guided dosing and find additional genetic association are necessary.
Figures and Tables
Table 1
Characteristics of the Study Population

Table 2
Effects of VKORC1 1173C>T and CYP2C9 Genotypes on Warfarin Stable Dose

| Genotype | n | Warfarin dose (mg/day) | p value |
|---|---|---|---|
| VKORC1 1173 | <0.05 | ||
| CC | 1 | 6.0 | |
| CT | 17 | 4.6±1.9 | |
| TT | 83 | 3.6±1.2 | |
| CYP2C9*3 | 0.080 | ||
| *1/*1 | 97 | 3.8±1.4 | |
| *1/*3 | 4 | 2.6±0.5 | |
| *3/*3 | 0 |
Table 3
Contribution of Individual Variables to the Algorithm
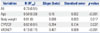
Table 4
Comparison of the Warfarin Dosing Algorithms

| Algorithm | Derivation sample (n) | R | R2 (R2adj) | Mean deviation (%) | Regression equation |
|---|---|---|---|---|---|
| This study | 101 | 0.73 | 0.53 (0.51) | 3.3 | y=0.406x+1.886 |
| Gage, et al.14 | 1015 | 0.57 | 0.33 (0.32) | -7.1 | y=0.453x+1.546 |
| Sconce, et al.15 | 297 | 0.62 | 0.38 (0.38) | -9.9 | y=0.394x+1.676 |
| Wu, et al.21 | 92 | 0.36 | 0.13 (0.12) | 46.4 | y=0.208x+4.105 |
| Anderson, et al.20 | 213 | 0.69 | 0.48 (0.47) | 4.3 | y=0.301x+2.427 |
| Ohno, et al.16 | 125 | 0.65 | 0.42 (0.41) | -10.0 | y=0.316x+1.913 |
| Huang, et al.19 | 266 | 0.49 | 0.24 (0.23) | -16.9 | y=0.237x+1.940 |
| Wadelius, et al.17 | 1496 | 0.63 | 0.40 (0.39) | 50.8 | y=0.360x+3.748 |
| IWPC13 | 4043 | 0.61 | 0.37 (0.37) | -17.9 | y=0.324x+1.634 |
| Cho, et al.18 | 130 | 0.64 | 0.41 (0.41) | -49.7 | y=0.094x+1.336 |
ACKNOWLEDGEMENTS
This study was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (A120030).
References
2. Whitlon DS, Sadowski JA, Suttie JW. Mechanism of coumarin action: significance of vitamin K epoxide reductase inhibition. Biochemistry. 1978; 17:1371–1377.

3. Stafford RS, Radley DC. The underutilization of cardiac medications of proven benefit, 1990 to 2002. J Am Coll Cardiol. 2003; 41:56–61.

4. Pirmohamed M. Warfarin: almost 60 years old and still causing problems. Br J Clin Pharmacol. 2006; 62:509–511.

5. Hylek EM, Singer DE. Risk factors for intracranial hemorrhage in outpatients taking warfarin. Ann Intern Med. 1994; 120:897–902.

6. Hylek EM, Skates SJ, Sheehan MA, Singer DE. An analysis of the lowest effective intensity of prophylactic anticoagulation for patients with nonrheumatic atrial fibrillation. N Engl J Med. 1996; 335:540–546.

7. Loebstein R, Yonath H, Peleg D, Almog S, Rotenberg M, Lubetsky A, et al. Interindividual variability in sensitivity to warfarin--nature or nurture? Clin Pharmacol Ther. 2001; 70:159–164.

8. Scordo MG, Caputi AP, D'Arrigo C, Fava G, Spina E. Allele and genotype frequencies of CYP2C9, CYP2C19 and CYP2D6 in an Italian population. Pharmacol Res. 2004; 50:195–200.

9. Bae JW, Kim HK, Kim JH, Yang SI, Kim MJ, Jang CG, et al. Allele and genotype frequencies of CYP2C9 in a Korean population. Br J Clin Pharmacol. 2005; 60:418–422.

10. Lindh JD, Holm L, Andersson ML, Rane A. Influence of CYP2C9 genotype on warfarin dose requirements--a systematic review and meta-analysis. Eur J Clin Pharmacol. 2009; 65:365–375.

11. Kim HS, Lee SS, Oh M, Jang YJ, Kim EY, Han IY, et al. Effect of CYP2C9 and VKORC1 genotypes on early-phase and steady-state warfarin dosing in Korean patients with mechanical heart valve replacement. Pharmacogenet Genomics. 2009; 19:103–112.

12. Yang L, Ge W, Yu F, Zhu H. Impact of VKORC1 gene polymorphism on interindividual and interethnic warfarin dosage requirement--a systematic review and meta analysis. Thromb Res. 2010; 125:e159–e166.
13. International Warfarin Pharmacogenetics Consortium. Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009; 360:753–764.

14. Gage BF, Eby C, Johnson JA, Deych E, Rieder MJ, Ridker PM, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008; 84:326–331.

15. Sconce EA, Khan TI, Wynne HA, Avery P, Monkhouse L, King BP, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005; 106:2329–2333.

16. Ohno M, Yamamoto A, Ono A, Miura G, Funamoto M, Takemoto Y, et al. Influence of clinical and genetic factors on warfarin dose requirements among Japanese patients. Eur J Clin Pharmacol. 2009; 65:1097–1103.

17. Wadelius M, Chen LY, Lindh JD, Eriksson N, Ghori MJ, Bumpstead S, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009; 113:784–792.

18. Cho HJ, On YK, Bang OY, Kim JW, Huh W, Ko JW, et al. Development and comparison of a warfarin-dosing algorithm for Korean patients with atrial fibrillation. Clin Ther. 2011; 33:1371–1380.

19. Huang SW, Chen HS, Wang XQ, Huang L, Xu DL, Hu XJ, et al. Validation of VKORC1 and CYP2C9 genotypes on interindividual warfarin maintenance dose: a prospective study in Chinese patients. Pharmacogenet Genomics. 2009; 19:226–234.

20. Anderson JL, Horne BD, Stevens SM, Grove AS, Barton S, Nicholas ZP, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007; 116:2563–2570.

21. Wu AH, Wang P, Smith A, Haller C, Drake K, Linder M, et al. Dosing algorithm for warfarin using CYP2C9 and VKORC1 genotyping from a multi-ethnic population: comparison with other equations. Pharmacogenomics. 2008; 9:169–178.

22. Yamaguchi T. Optimal intensity of warfarin therapy for secondary prevention of stroke in patients with nonvalvular atrial fibrillation: a multicenter, prospective, randomized trial. Japanese Nonvalvular Atrial Fibrillation-Embolism Secondary Prevention Cooperative Study Group. Stroke. 2000; 31:817–821.

23. Zhou SF, Wang B, Yang LP, Liu JP. Structure, function, regulation and polymorphism and the clinical significance of human cytochrome P450 1A2. Drug Metab Rev. 2010; 42:268–354.

24. Holbrook AM, Pereira JA, Labiris R, McDonald H, Douketis JD, Crowther M, et al. Systematic overview of warfarin and its drug and food interactions. Arch Intern Med. 2005; 165:1095–1106.

25. Limdi NA, Wadelius M, Cavallari L, Eriksson N, Crawford DC, Lee MT, et al. Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood. 2010; 115:3827–3834.

26. Zhu Y, Shennan M, Reynolds KK, Johnson NA, Herrnberger MR, Valdes R Jr, et al. Estimation of warfarin maintenance dose based on VKORC1 (-1639 G>A) and CYP2C9 genotypes. Clin Chem. 2007; 53:1199–1205.

27. Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther. 2008; 83:460–470.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download