Abstract
Over the past three decades, a large number of genetic studies have been aimed at finding genetic variants associated with the risk of asthma, applying various genetic and genomic approaches including linkage analysis, candidate gene polymorphism studies, and genome-wide association studies (GWAS). However, contrary to general expectation, even single nucleotide polymorphisms (SNPs) discovered by GWAS failed to fully explain the heritability of asthma. Thus, application of rare allele polymorphisms in well defined phenotypes and clarification of environmental factors have been suggested to overcome the problem of 'missing' heritability. Such factors include allergens, cigarette smoke, air pollutants, and infectious agents during pre- and post-natal periods. The first and simplest interaction between a gene and the environment is a candidate interaction of both a well known gene and environmental factor in a direct physical or chemical interaction such as between CD14 and endotoxin or between HLA and allergens. Several GWAS have found environmental interactions with occupational asthma, aspirin exacerbated respiratory disease, tobacco smoke-related airway dysfunction, and farm-related atopic diseases. As one of the mechanisms behind gene-environment interaction is epigenetics, a few studies on DNA CpG methylation have been reported on subphenotypes of asthma, pitching the exciting idea that it may be possible to intervene at the junction between the genome and the environment. Epigenetic studies are starting to include data from clinical samples, which will make them another powerful tool for research on gene-environment interactions in asthma.
Asthma is a genetically complex disease associated with familial segregation of atopy and increased levels of total serum IgE.1 Asthma and atopy are closely correlated with increased bronchial hyperresponsiveness2 and elevated blood eosinophil count.3 These intermediate phenotypes are highly heritable and subject to intensive genetic research. Asthmatics cluster in families, indicating that a genetic component is likely to be involved. Twin studies represent a useful first step to determine whether a given trait or disease has a measurable genetic component. In a large twin study with 7000 same-sex twins born between 1886 and 1925, the concordance rate for self-reported asthma in monozygotic (MZ) twin pairs was 19%, four times as high as the 4.8% in dizygotic (DZ) twins.4 Heritability has been estimated as up to 60% and is thought to be determined by genetic factors such as nucleotide variants.
To search for nucleotide variants as genetic factors, genome-wide linkage analyses, biologically plausible candidate-gene approaches, and genome-wide association studies (GWAS) have been used over the past 30 years. While monogenic disorders with simple Mendelian inheritance such as cystic fibrosis have been successfully identified using whole genome linkage analysis, positional cloning, and case control studies,5 more than 100 loci on autosomal and sex chromosomes have been found to be linked to asthma. Among them, at least 5 asthma genes, including a disintegrin and metalloprotease 33 (ADAM33) on 20p13,6 dipeptidyl peptidase 10 (DPP10) on 2q14.1,7 plant homeodomain zinc finger protein 11 (PHF11) on 13q14.2,8 G protein-coupled receptor (GPR) for asthma susceptibility on 7p15-p14,9 and prostaglandin D2 receptor (PTGDR) on 14q24,10 have been identified as strongly associated with asthma. However, replication studies including a large cohort of 7703 adults revealed that only a minor risk of increase in asthma incidence was linked to DPP10 and ADAM33.6,7 These limitations of linkage analysis in complex human disease have caused a redirection of focus from linkage analysis and microsatellite markers toward single nucleotide polymorphisms (SNP) genotyping and different analytical strategies based on association and haplotype analysis.11
Consequently, the candidate gene approach led to more than 300 genes containing asthma-associated SNPs recorded in the NCBI databases (www.ncbi.nlm.nih.gov). As of March 2015, a keyword search for "asthma and polymorphism" returned 2851 publications, including 226 papers published in Korea. Interestingly, almost all associated SNPs have odds ratios (ORs) below 2.0, indicating that the candidate gene approach has provided information on genetic variants causing a significant risk of increase in asthma, but their contribution to the development of asthma may be smaller than expected.
Since the development of GWAS, genotyping of 500000 SNPs enables nearly complete surveys of all common genetic variability.12 Based on this concept, whole genome SNP genotyping arrays have been developed and employed for investigating the genetic background of multifactorial complex diseases over the past 8 years. As of February 2012, 2111 GWAS had been published showing statistical significance for 17 traits of common allele complex diseases (NHGRI catalogue at www.genome.gov/gwastudies). GWAS on asthma and its traits have been published from 2007 onwards, producing 408 publication titles containing "GWAS and Asthma" in PubMed, including 23 with Korean asthmatics. Among the initial eight studies (Table 1), six pertained to Caucasians, one to Mexicans, and Koreans were included in two studies. Seven studies focused on asthma risk, the remaining one on occupational asthma.
The first GWAS on asthma, published in England in 2007,13 found strong signals on chromosome 17q and on chromosome 2. SNPs associated with childhood asthma could be consistently linked to transcript levels of ORMDL3, a member of a gene family that encodes transmembrane proteins anchored in the endoplasmic reticulum.13 The second asthma GWAS analyzed sequence variants affecting blood eosinophil counts in Icelanders, and the ten most significant SNPs were further studied in 12118 Europeans and 5212 East Asians including Koreans. While SNP rs1420101 in the IL1RL1/IL18R1 gene cluster at 2q12 was strongly associated with asthma (p=5.5×10-12), its OR was still below 1.5. In 2010, a large-scale, consortium-based GWAS of asthma with 10365 asthmatics and 16110 controls confirmed the association of the previously defined SNPs including the ones in ORMDL3 and IL1RL1,14 yet their ORs were within the range of 0.5 to 1.5.
Subsequently, GWAS has been applied to determine whether disease-related intermediate phenotypes are causal or secondary to the disease progress.15 A meta-analysis on total IgE concentration showed strong association with HLA-DQ (rs9469220) in Caucasians16 and CRIM1 in Koreans (rs848512 and rs711254).17
GWAS have identified hundreds of genetic variants associated with complex human diseases and traits, providing deep insights into their genetic architecture. In case of age-related macular degeneration, proportions of heritability explained by 5 SNPs were found to be responsible for 50% of the risk (Table 2), while for Crohn's disease 32 loci can explain 20% of the heritability. However, most variants identified so far confer relatively small increments in risk, explaining only a small proportion of heritability and leading to the question how the missing heritability can be explained.18 In the GWAS presented in Table 1, the population attributable risk fractions (PAF) of SNPs in asthma and its' traits ranged from 3.9% to 24%, indicating a relatively large impact of the genetic variants on asthma development. In the studies including asthmatics, rs7216389 in ORMDL3 returned a PAF of 21.8%, while the SNPs on the other gene had a PAF of less than 12%. This suggests that even the GWAS-discovered SNPs explain only limited genetic effects on the development of asthma.
Considering the causative factors of GWAS' limitations such as imprecise disease phenotypes, use of control groups of questionable comparability, and inconsideration of environmental contributors, Manolio, et al.18 proposed the following three solutions to overcome the missing heritability: discovery of SNPs of rare allele frequencies, stratification of subjects into well defined specific phenotypes, and clarification of environmental influences.
Until recently, much of the speculation about missing heritability from GWAS has focused on the possible contribution of rare variants (minor allele frequency MAF<0.5%), because the previous GWAS had analyzed common variants of MAF>5%. To provide more information on these rare variants on human chromosomes, the 1000 Genomes Project (www.1000genomes.org/page.php) of sequencing 1000 individual genomes has already identified more than 15 million new SNPs, 1 million short insertions and deletions, and 20000 structural variants.19 The Project pilot data revealed that African populations show the highest density of rare variants followed by Asian and European populations.20 The newly discovered SNPs have started to be applied to recent genetic association studies on the traits of asthma.21
The definition of phenotype under study is of major importance, as asthma is a heterogeneous disease. Well defined subphenotypes improve the genetic power of SNPs. The ORMDL3 SNP was reanalyzed in Caucasians and Koreans according to onset age of asthma development (Table 3).22 When subjects were stratified around age 16, the association of rs7216389 at 17q21 became more apparent in Caucasians and Koreans under that age, with ODs ranging from 1.26 to 1.49, while ODs of the older group ranged from 0.87 to 1.12 (Fig. 1). Attributable risk fractions were 0.269 in the early and 0.057 in the late onset group. These data indicate the necessity of stratification of asthma phenotypes according to age, because the two types of asthma may have different pathogeneses.
Various ways have been tried to develop subphenotypes. Schematically, the pathogenic mechanism of asthma can be divided into two major pathways23-the adaptive immune response pathway and the innate immunity pathway. In the former, Th2 cells, mast cells, and eosinophils participate in an antigen-specific IgE- and Th2 cytokine-dependent manner, a process usually starting at young age. In the latter, macrophages, dendritic cells, epithelial cells, and neutrophils are involved, causing IgE-independent, adult onset asthma. Therefore, asthmatics should be stratified into several subphenotypes as shown in Table 4.
Regarding environmental and lifestyle factors as triggers, asthma can be subgrouped into IgE-dependent allergic asthma, aspirin-exacerbated respiratory disease, occupational asthma, exercise-induced asthma, and menstruation- or obesity-associated asthma. Inflammatory patterns on sputum analysis have revealed eosinophilic, neutrophilic, and paucigranulocytic types of asthma. Therefore, asthmatics can be stratified into several clinical and physiological subgroups. Recently, biological phenotypes (endotypes) have been introduced to classify parts of the asthma process. Many biological mediators such as immune and constitutional cells in allergy, inflammation, and airway remodeling are candidates for determining endotypes, e.g., Th2, Th17, innate Th2, epithelial and smooth muscle dysfunctions (Table 4). Thus, future genetic association studies should be applied to asthmatics stratified into well defined subphenotypes using cluster analysis and new endotypes.24,25
Whereas there are certainly genetic factors involved in the development of multifactorial diseases, the prevalence of childhood and adult-onset asthma has increased dramatically in both developed and developing countries during the last two to three decades.26 Epidemiological studies have demonstrated that the entirety of SNPs discovered so far is not able to explain all phenotypic differences. The twin cohort study mentioned above, for example, showed a concordance rate for self-reported asthma in MZ twin pairs of 19%.27 Assuming that MZ twins have identical genetic variants, they should develop asthma almost concurrently. However, a concordance rate of less than 20% indicates that non-genetic factors may play a role in the development of asthma. One possible explanation for this discordance is epigenetics, which is fundamentally affected by the environment. Although the concept of gene-environment (GxE) interactions emerged in the 19th century, investigation of GxE interactions has been mainly limited to candidate genes or candidate environmental exposures. In 1938, Haldane28 suggested that genetic differences might explain the variability of respiratory symptoms and survival of potters in response to industrial exposures.
While the effects of environmental factors certainly depend on genetic susceptibility, exposure to smoking and malnutrition during pre- and post-natal periods are well known risk factors for asthma. Moreover, asthma is triggered and exacerbated by allergens,29,30,31,32 indoor and outdoor air pollutants,33 occupational exposures,34,35 microbial and viral pathogens,36,37,38 nutrition,39,40 and lifestyle.41,42
The first and simplest situation is a candidate interaction: both the gene and the environmental factor are known and assumed to be involved in a kind of direct physical or chemical reaction. Well known and extensively studied candidate interactions are the interactions between the CD14 gene and environmental exposure to endotoxin, an essential component of the gram-negative bacterial cell wall,43,44 between Toll-like receptor genes and infectious agents,45 and between HLA genes and allergens,46 which are presented to T lymphocytes via HLA molecules on dendritic cells.
Occupational exposure to inducers or triggers is an important and easily accessible environmental factor, e.g., toluene diisocyanate (TDI)-induced asthma in Korean patients as analyzed by GWAS.47 Genetic polymorphisms of CTNNA3 (catenin alpha 3, alpha-T catenin) were significantly associated (OR=5.84 for rs10762058) with the TDI-induced asthma phenotype, and PAF increased up to 24%, indicating that missing heritability of asthma can be compensated by introduction of environment factors into the genetic analysis. Other examples of environmental effects on susceptible individuals are drug-induced reactions such as aspirin exacerbated respiratory disease (AERD). Association studies on AERD began with biologically plausible genes responsible for over- or under-production of critical modulators in the metabolism of arachidonic acids. LTC4S, ALOX5, NAT2, CysLTR1, and CYSLTR2 of the cysteinyl leukotriene pathway have AERD-associated SNPs with ORs ranging from 1.88 to 9.78 (Fig. 2).48 Mediators in the pathways of lipoxins, thromboxanes, and prostaglandins also contribute to adverse aspirin reactions, with some genes of those pathways, and others discovered by GWAS,49 showing strong, yet slightly lower ORs than the genes of the cysteinyl leukotriene pathway. However, diagnosis of AERD is hampered by the low penetrance of these genotypes, i.e., if genetically susceptible subjects do not take aspirin or nonsteroidal anti-inflammatory drugs (NSAID) in their lifetime, they will not develop AERD or aspirin-induced urticaria. Thus, history of aspirin or NSAID intake should be assessed at diagnosis, although exact amounts and times of intake are hardly recorded by patients or physicians.
In the first asthma GWAS, identifying a strong association of asthma with the 17q21 locus (Fig. 3),48 a further interesting finding on 17q21 variants came out after the 1511 subjects from 372 families were grouped by passive exposure to environmental tobacco smoke early in life.50 ORMDL3 variants at rs8076131 showed significant association (OR=2.5) with the risk of asthma in families with offspring exposed to cigarette smoke, in contrast to the unexposed (OR=1.38). This result, which was later replicated,51,52 hinted at the importance of environmental factors in genetic studies on asthma (Fig. 4). The first published genome-wide GxE interaction study for asthma explored interactions between farm-related exposure and genome-wide SNPs in 1700 children from four rural areas in Central Europe.53 This well designed study did not reveal any significant interactions with common SNPs previously associated with asthma. Nevertheless, interactions were detected with rare SNPs, e.g., in genes of the glutamate receptor pathway.54 For the most complex future scenario in which both genes and environment are unknown, exposomes-large sets of environmental exposures55-and specific asthma phenotypes56 need to be defined. While agnostic approaches to GxE interactions are still in their infancy, methodological progress towards optimization of study design and analytical methods has been made.57,58 Recently, both gut and airway microbiomes were found to contribute to chronic asthma,59 introducing environmental microbiomes as further complicating factors of complex GxE interactions in asthma development.
Epigenetics comprises changes in gene expression due to mechanisms other than variations of the underlying DNA sequence,60 which may induce phenotypic alterations and persist through cell divisions for the remainder of a cell's life, possibly lasting for multiple generations without any change in the intrinsic DNA sequence of an organism. Epigenetic changes include histone deacetylation, DNA methylation, and non-coding RNAs. Particularly regions of methylated DNA, i.e., methyl groups covalently added to cytosine residues in CpG dinucleotides,61 have been correlated with tissue-specific expression of several genes and with active coding regions across the genome.62
In the human genome, about one fifth of CpG islands at 5' UTRs are variably methylated, and one third of methylations correlate with amounts of transcripts. As epigenetics plays a role in main immunological aspects of asthma such as T-cell differentiation and regulation, genome-wide studies of methylation status at various loci may identify new asthma initiating and modulating genes and pathways.
So far, genome-wide CpG methylation changes have been found in a few studies of asthmatics, e.g., methylation patterns of whole genome DNA were significantly different between nasal polyps of subjects with AERD and aspirin tolerant asthma (ATA) patients, whereas less differences were found in buffy coat.63 Differences between AERD and ATA comprised 332 CpG sites in 296 genes that were hypomethylated, and 158 sites in 141 genes that were hypermethylated (Fig. 5). CpG site methylations of nasal polyps did not correlate with those of buffy coats, indicating that differences in methylation patterns were a nasal tissue-specific finding. Among the genes of the arachidonic acid pathway, prostaglandin E synthase was hypermethylated and prostaglandin D synthase, arachidonate 5-lipoxygenase-activating protein, leukotriene B4 receptor, and lipoxygenase homology domains 1 were hypomethylated, indicating that different methylation patterns of these candidate genes may be responsible for the penetrance of specific phenotypes such as AERD in asthma. In the bronchial mucosa of atopic asthmatics, hypermethylation was detected at 6 loci in 6 genes, while hypomethylation was found at 49 loci in 48 genes, compared to non-atopic asthmatics.64
The definition of phenotypes is of major importance to genetic studies. Ways of improving phenotype definition include physiologic or biological phenotypes related to disease processes (endotypes) and the use of unbiased and statistically based approaches such as cluster analysis, which are promising because they may define new phenotypes from a number of features related to disease occurrence, evolution, therapy, and biological phenotypes. As time-dependent change plays a major role in clustering-based approaches, more precise phenotyping should be obtainable from longitudinal studies. Taking into account time of asthma onset can characterize novel GxE interactions, extending the two-dimensional problem of GxE interactions to a three-dimensional problem. Although most genetic studies so far have examined single phenotypes, joint analysis of multiple phenotypes through multidimensional and multivariate methods can improve statistical power for identifying genes with a pleiotropic effect. Although a number of multivariate methods have been proposed, improvements are needed to integrate information from various data types, including "-omics" data. Moreover, as for GWAS, meta-analysis of gene-environment interaction studies can increase driving power further and provide robust estimates of GxE interactions. As the identification of functional regulatory elements for human gene expression is an active subject of research in the postgenomic era, longitudinal studies, including collection of biological samples over time, are a desirable way to validate the positive results obtained on GxE interactions.
Figures and Tables
 | Fig. 1Age-dependent changes of odds ratios of T allele rs7216389 in ORMDL3 in respect to the association with asthma. Permission license number: 3596250009660. Taken with permission from Halapi, et al. Eur J Hum Genet 2010;18:902-8.22
|
 | Fig. 2Genetic impact of SNPs in candidate genes of the arachidonic acid pathway on aspirin exacerbated respiratory disease. Data are represented as odds ratios (OR). Taken with permission from Park, et al. Allergy Asthma Immunol Res 2013;5:258-76.48 SNPs, single nucleotide polymorphisms. |
 | Fig. 3Genetic impact of SNPs in candidate genes of immune and inflammatory pathways on aspirin exacerbated respiratory disease. Data are represented as odds ratios (OR). Taken with permission from Park, et al. Allergy Asthma Immunol Res 2013;5:258-76.48 SNPs, single nucleotide polymorphisms. |
 | Fig. 4Example of candidate interaction between gene and environment. A variant (rs8076131) in ORMDL3 (gene) is associated with early asthma and exposure to environmental tobacco smoke (ETS, environment). OR increases two times in cases with positive ETS than in cases with negative ETS. Modified from Bouzigon, et al.50 OR, odds ratio. |
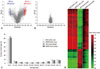 | Fig. 5CpG DNA methylation patterns of nasal polyps and peripheral blood mononuclear cells obtained from subjects with AERD and ATA. (A) Volcano plot of differential methylation levels between AERD and ATA in nasal polyp tissues (A) and buffy coat samples (B). Red dots, deltabeta≥0.5 and p value≤0.01; blue dots, deltabeta≤-0.5 and p value≤0.01; grey dots, -0.5≤deltabeta≤0.5 and p value>0.01. Deltabeta, difference of DNA methylation level (subtracting DNA methylation level of ATA from AERD). -log (p), log-transformed t-test p values. (C) Distribution of DNA methylation levels of AERD and ATA in buffy coat and nasal polyps. Average Beta, DNA methylation level (0 to 1). (D) Heat map of 490 differentially methylated CpGs between AERD and ATA in buffy coat and nasal polyps. Taken with permission from Park, et al. Allergy Asthma Immunol Res 2013;5:258-76.48 AERD, aspirin exacerbated respiratory disease; ATA, aspirin tolerant asthma. |
Table 1
Genes Associated with Asthma by GWAS Studies between 2007-2010

| No. | Gene | Ethnicities | OR | References |
|---|---|---|---|---|
| 1 | ORMDL3 | Caucasian | 1.45 | Moffatt, et al.13 |
| 2 | CTNNA3 | Korean | 1.85 | Kim, et al.47 |
| 3 | IL1RL1 | Caucasian/Korean/Taiwan | 1.16 | Gudbjartsson, et al.65 |
| 4 | PDE4D | Caucasian | 2.32 | Himes, et al.66 |
| 5 | TLE4 | Mexican | 1.68 | Mashimo, et al.67 |
| 6 | DENND1B | Caucasian | 1.83 | Sleiman, et al.68 |
| 7 | RAD50-IL13 | Caucasian | 1.64 | Li, et al.69 |
| 8 | IL1RL1/IL18R1 | Caucasian | <1.5 | Moffatt, et al.14 |
| ORMDL3 |
Table 2
Estimates of Heritability and Number of Loci for Several Complex Traits

HDL, high density lipoprotein.
*Residual is after adjustment for age, gender, diabetes. Adapted from Manolio, et al.18
Table 3
Comparison of Odds Ratios of T Allele rs7216389 in ORMDL3 between Early Onset and Adult Onset of Asthma
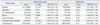
na/nc, number of asthmatics and controls; OR, odds ratio; CI, confidence intervals.
Adapted from Halapi, et al.22
Table 4
Categories of Asthma According to Etiology, Clinical Patterns, and Underlying Mechanisms

ACKNOWLEDGEMENTS
The study was supported by Soonchunhyang University research fund and by the Ministry of Health and Welfare, Republic of Korea (HI13C0319).
References
1. Burrows B, Martinez FD, Halonen M, Barbee RA, Cline MG. Association of asthma with serum IgE levels and skin-test reactivity to allergens. N Engl J Med. 1989; 320:271–277.

2. Burrows B, Sears MR, Flannery EM, Herbison GP, Holdaway MD, Silva PA. Relation of the course of bronchial responsiveness from age 9 to age 15 to allergy. Am J Respir Crit Care Med. 1995; 152(4 Pt 1):1302–1308.

3. Bousquet J, Chanez P, Vignola AM, Lacoste JY, Michel FB. Eosinophil inflammation in asthma. Am J Respir Crit Care Med. 1994; 150(5 Pt 2):S33–S38.

5. Zielenski J, Tsui LC. Cystic fibrosis: genotypic and phenotypic variations. Annu Rev Genet. 1995; 29:777–807.

6. Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002; 418:426–430.

7. Allen M, Heinzmann A, Noguchi E, Abecasis G, Broxholme J, Ponting CP, et al. Positional cloning of a novel gene influencing asthma from chromosome 2q14. Nat Genet. 2003; 35:258–263.

8. Zhang Y, Leaves NI, Anderson GG, Ponting CP, Broxholme J, Holt R, et al. Positional cloning of a quantitative trait locus on chromosome 13q14 that influences immunoglobulin E levels and asthma. Nat Genet. 2003; 34:181–186.

9. Laitinen T, Polvi A, Rydman P, Vendelin J, Pulkkinen V, Salmikangas P, et al. Characterization of a common susceptibility locus for asthma-related traits. Science. 2004; 304:300–304.

10. Oguma T, Palmer LJ, Birben E, Sonna LA, Asano K, Lilly CM. Role of prostanoid DP receptor variants in susceptibility to asthma. N Engl J Med. 2004; 351:1752–1763.

12. Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009; 360:1759–1768.

13. Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007; 448:470–473.

14. Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010; 363:1211–1221.

15. Sheehan NA, Didelez V, Burton PR, Tobin MD. Mendelian randomisation and causal inference in observational epidemiology. PLoS Med. 2008; 5:e177.

16. Levin AM, Mathias RA, Huang L, Roth LA, Daley D, Myers RA, et al. A meta-analysis of genome-wide association studies for serum total IgE in diverse study populations. J Allergy Clin Immunol. 2013; 131:1176–1184.

17. Kim JH, Cheong HS, Park JS, Jang AS, Uh ST, Kim YH, et al. A genome-wide association study of total serum and mite-specific IgEs in asthma patients. PLoS One. 2013; 8:e71958.

18. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009; 461:747–753.

19. 1000 Genomes Project Consortium. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010; 467:1061–1073.

20. Raska P, Zhu X. Rare variant density across the genome and across populations. BMC Proc. 2011; 5:Suppl 9. S39.

21. Ortega VE, Hawkins GA, Moore WC, Hastie AT, Ampleford EJ, Busse WW, et al. Effect of rare variants in ADRB2 on risk of severe exacerbations and symptom control during longacting β agonist treatment in a multiethnic asthma population: a genetic study. Lancet Respir Med. 2014; 2:204–213.

22. Halapi E, Gudbjartsson DF, Jonsdottir GM, Bjornsdottir US, Thorleifsson G, Helgadottir H, et al. A sequence variant on 17q21 is associated with age at onset and severity of asthma. Eur J Hum Genet. 2010; 18:902–908.

23. Green RH, Brightling CE, Bradding P. The reclassification of asthma based on subphenotypes. Curr Opin Allergy Clin Immunol. 2007; 7:43–50.

24. Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010; 181:315–323.

25. Jang AS, Kwon HS, Cho YS, Bae YJ, Kim TB, Park JS, et al. Identification of subtypes of refractory asthma in Korean patients by cluster analysis. Lung. 2013; 191:87–93.

26. Ho SM. Environmental epigenetics of asthma: an update. J Allergy Clin Immunol. 2010; 126:453–465.

27. Thomsen SF, van der Sluis S, Kyvik KO, Skytthe A, Backer V. Estimates of asthma heritability in a large twin sample. Clin Exp Allergy. 2010; 40:1054–1061.

28. Haldane JBS. Heredity and politics. London: George Allen & Unwin Ltd Pblishe;1938.
29. Huiyan W, Yuhe G, Juan W, Junyan Z, Shan W, Xiaojun Z, et al. The Importance of Allergen Avoidance in High Risk Infants and Sensitized Patients: A Meta-analysis Study. Allergy Asthma Immunol Res. 2014; 6:525–534.

30. Peden D, Reed CE. Environmental and occupational allergies. J Allergy Clin Immunol. 2010; 125:2 Suppl 2. S150–S160.

32. Park HJ, Lim HS, Park KH, Lee JH, Park JW, Hong CS. Changes in allergen sensitization over the last 30 years in Korea respiratory allergic patients: a single-center. Allergy Asthma Immunol Res. 2014; 6:434–443.

33. Lee JH, Lee HS, Park MR, Lee SW, Kim EH, Cho JB, et al. Relationship between indoor air pollutant levels and residential environment in children with atopic dermatitis. Allergy Asthma Immunol Res. 2014; 6:517–524.

34. Jeong I, Kim I, Park HJ, Roh J, Park JW, Lee JH. Allergic diseases and multiple chemical sensitivity in Korean adults. Allergy Asthma Immunol Res. 2014; 6:409–414.

35. Hur GY, Ye YM, Koh DH, Kim SH, Park HS. IL-4 Receptor α Polymorphisms May Be a Susceptible Factor for Work-Related Respiratory Symptoms in Bakery Workers. Allergy Asthma Immunol Res. 2013; 5:371–376.

36. Yang SN, Hsieh CC, Kuo HF, Lee MS, Huang MY, Kuo CH, et al. The effects of environmental toxins on allergic inflammation. Allergy Asthma Immunol Res. 2014; 6:478–484.

37. Kim BJ, Lee SY, Kim HB, Lee E, Hong SJ. Environmental changes, microbiota, and allergic diseases. Allergy Asthma Immunol Res. 2014; 6:389–400.

38. Yang SI, Lee E, Jung YH, Kim HY, Seo JH, Kwon JW, et al. Effect of antibiotic use and mold exposure in infancy on allergic rhinitis in susceptible adolescents. Ann Allergy Asthma Immunol. 2014; 113:160–165.e1.

39. Kim SH, Sutherland ER, Gelfand EW. Is there a link between obesity and asthma? Allergy Asthma Immunol Res. 2014; 6:189–195.

40. Lee SY, Kang MJ, Kwon JW, Park KS, Hong SJ. Breastfeeding Might Have Protective Effects on Atopy in Children With the CD14C-159T CT/CC Genotype. Allergy Asthma Immunol Res. 2013; 5:239–241.

41. Seo S, Kim D, Paul C, Yoo Y, Choung JT. Exploring Household-level Risk Factors for Self-reported Prevalence of Allergic Diseases Among Low-income Households in Seoul, Korea. Allergy Asthma Immunol Res. 2014; 6:421–427.

42. Zhang G, Khoo SK, Mäkelä MJ, Candelaria P, Hayden CM, von Hertzen L, et al. Maternal Genetic Variants of IL4/IL13 Pathway Genes on IgE With "Western or Eastern Environments/Lifestyles". Allergy Asthma Immunol Res. 2014; 6:350–356.

43. Vercelli D. Learning from discrepancies: CD14 polymorphisms, atopy and the endotoxin switch. Clin Exp Allergy. 2003; 33:153–155.

44. Martinez FD. CD14, endotoxin, and asthma risk: actions and interactions. Proc Am Thorac Soc. 2007; 4:221–225.

45. Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000; 406:782–787.

47. Kim SH, Cho BY, Park CS, Shin ES, Cho EY, Yang EM, et al. Alpha-T-catenin (CTNNA3) gene was identified as a risk variant for toluene diisocyanate-induced asthma by genome-wide association analysis. Clin Exp Allergy. 2009; 39:203–212.

48. Park SM, Park JS, Park HS, Park CS. Unraveling the genetic basis of aspirin hypersensitivity in asthma beyond arachidonate pathways. Allergy Asthma Immunol Res. 2013; 5:258–276.

49. Park BL, Kim TH, Kim JH, Bae JS, Pasaje CF, Cheong HS, et al. Genome-wide association study of aspirin-exacerbated respiratory disease in a Korean population. Hum Genet. 2013; 132:313–321.

50. Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008; 359:1985–1994.

51. Flory JH, Sleiman PM, Christie JD, Annaiah K, Bradfield J, Kim CE, et al. 17q12-21 variants interact with smoke exposure as a risk factor for pediatric asthma but are equally associated with early-onset versus late-onset asthma in North Americans of European ancestry. J Allergy Clin Immunol. 2009; 124:605–607.

52. van der Valk RJ, Duijts L, Kerkhof M, Willemsen SP, Hofman A, Moll HA, et al. Interaction of a 17q12 variant with both fetal and infant smoke exposure in the development of childhood asthma-like symptoms. Allergy. 2012; 67:767–774.

53. Ege MJ, Strachan DP, Cookson WO, Moffatt MF, Gut I, Lathrop M, et al. Gene-environment interaction for childhood asthma and exposure to farming in Central Europe. J Allergy Clin Immunol. 2011; 127:138–144. 144.e1-4.

54. Hamza TH, Chen H, Hill-Burns EM, Rhodes SL, Montimurro J, Kay DM, et al. Genome-wide gene-environment study identifies glutamate receptor gene GRIN2A as a Parkinson's disease modifier gene via interaction with coffee. PLoS Genet. 2011; 7:e1002237.

56. Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012; 18:716–725.

57. Bookman EB, McAllister K, Gillanders E, Wanke K, Balshaw D, Rutter J, et al. Gene-environment interplay in common complex diseases: forging an integrative model-recommendations from an NIH workshop. Genet Epidemiol. 2011; 35:217–225.

58. Thomas D. Gene--environment-wide association studies: emerging approaches. Nat Rev Genet. 2010; 11:259–272.

59. Han MK, Huang YJ, Lipuma JJ, Boushey HA, Boucher RC, Cookson WO, et al. Significance of the microbiome in obstructive lung disease. Thorax. 2012; 67:456–463.

60. Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007; 128:635–638.

61. Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006; 31:89–97.

62. Sato S, Yagi S, Arai Y, Hirabayashi K, Hattori N, Iwatani M, et al. Genome-wide DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) residing in mouse pluripotent stem cells. Genes Cells. 2010; 15:607–618.

63. Cheong HS, Park SM, Kim MO, Park JS, Lee JY, Byun JY, et al. Genome-wide methylation profile of nasal polyps: relation to aspirin hypersensitivity in asthmatics. Allergy. 2011; 66:637–644.

64. Kim YJ, Park SW, Kim TH, Park JS, Cheong HS, Shin HD, et al. Genome-wide methylation profiling of the bronchial mucosa of asthmatics: relationship to atopy. BMC Med Genet. 2013; 14:39.

65. Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009; 41:342–347.
66. Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009; 84:581–593.

67. Mashimo T, Hadjebi O, Amair-Pinedo F, Tsurumi T, Langa F, Serikawa T, et al. Progressive Purkinje cell degeneration in tambaleante mutant mice is a consequence of a missense mutation in HERC1 E3 ubiquitin ligase. PLoS Genet. 2009; 5:e1000784.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download