Abstract
Purpose
The role of IL28B gene variants and expression in hepatitis B virus (HBV) infections are not well understood. Here, we evaluated whether IL28B gene expression and rs12979860 variations are associated with HBV outcomes.
Materials and Methods
IL28B genetic variations (rs12979860) were genotyped by pyrosequencing of DNA samples from 137 individuals with chronic HBV infection [50 inactive carriers (IC), 34 chronic hepatitis B (CHB), 27 cirrhosis, 26 hepatocellular carcinoma (HCC)], and 19 healthy controls. IL28A/B mRNA expression in peripheral blood mononuclear cells was determined by qRT-PCR, and serum IL28B protein was measured by ELISA.
Results
Patients with IL28B C/C genotype had greater IL28A/B mRNA expression and higher IL28B protein levels than C/T patients. Within the various disease stages, compared to IC and healthy controls, IL28B expression was reduced in the CHB, cirrhosis, and HCC cohorts (CHB vs. IC, p=0.02; cirrhosis vs. IC, p=0.01; HCC vs. IC, p=0.001; CHB vs. controls, p<0.01; cirrhosis vs. controls, p<0.01; HCC vs. controls, p<0.01). When stratified with respect to serum HBV markers in the IC and CHB cohorts, IL28B mRNA and protein levels were higher in HBeAg-positive than negative individuals (p=0.01). Logistic regression analysis revealed that factors associated with high IL28B protein levels were C/C versus C/T genotype [p=0.016, odds ratio (OR)=0.25, 95% confidence interval (CI)=0.08-0.78], high alanine aminotransferase values (p<0.001, OR=8.02, 95% CI=2.64-24.4), and the IC stage of HBV infection (p<0.001).
Hepatitis B virus (HBV) infection is the most common infectious liver disease, affecting 350 million people worldwide.1,2 Persistent HBV infection often progresses from an inactive carrier state, to chronic hepatitis, then cirrhosis, and finally, hepatocellular carcinoma (HCC).3,4,5 This progression is the result of complicated host-virus interactions, with the host immune status playing an important role. While a host can clear the virus with an appropriate immune response, the virus can also avoid immune clearance through co-evolution. Attempts to clear chronic HBV infection are responsible for immune-mediated liver damage. As the patient ages, the immune response of the patient changes and typically becomes less effective with advancing age. This age-dependent change is also related to the genetic predisposition of the host.6,7 Robust genetic evidence from genome-wide association studies suggests that host genetic variations contribute significantly to the outcome of chronic HBV infections.8,9,10
Recently, interleukin 28B (IL28B) rs12979860 genetic variations have been associated with responses to interferon-α therapy and spontaneous viral clearance of the hepatitis C virus (HCV). Indeed, in two independent studies, IL28B polymorphisms were identified as the strongest predictor of treatment outcome.11,12 In terms of spontaneous clearance, Thomas, et al.13 analyzed the relationship between spontaneous HCV clearance and IL28B rs12979860 polymorphisms, and found that the C/C genotype was associated with enhanced clearance rates. In addition, Tanaka, et al.14 showed that HCV patients with G/G or G/T genotypes at IL28B rs8099917 had lower IL28 mRNA expression than T/T genotype patients. This finding may reflect the fact that different alleles of IL28B influence its expression at the transcriptional and protein levels.15 However, work by Urban, et al.16 showed no association between polymorphism and the expression of IL28B protein in the liver.
The role which IL28B gene variants and expression might play in influencing the course of chronic HBV infection remains unclear. Sonneveld, et al.17 reported that in patients with HBeAg-positive chronic HBV, IL28B polymorphisms were associated with HBeAg seroconversion and hepatitis B surface antigen (HBsAg) reduction in response to pegylated interferon alpha (PEG-IFNα) treatment. On the other hand, Peng, et al.18 reported that the IL28B rs12979860 polymorphism does not have a strong impact on HBV outcomes. Interestingly, Li, et al.19 found that serum IL28B protein levels were lower in chronic HBV patients, than in individuals with self-limited infection and healthy controls. However, these authors also showed that chronic HBV-infected patients with the C/C genotype have higher serum IL28B protein levels than patients with non-C/C genotypes.19 Recently, Noureddin, et al.20 found that IL28B C/C was associated with fibrosis progression and clinical outcomes in patients with chronic hepatitis C.
Thus, in an effort to more fully elucidate the role of IL28B variants in the natural history of HBV infection, we evaluated the relationship of IL28B polymorphisms and expression with the outcomes of chronic HBV in a Han Chinese cohort consisting of patients at different stages of chronic HBV infection.
Participating subjects with HBsAg-confirmed HBV infection were from the outpatient and inpatient pools at the Department of Gastroenterology, First Affiliated Hospital of Jilin University from November 2009 to June 2010. A total of 137 chronically infected patients and 19 HBsAg-negative, healthy volunteers provided informed consent and were enrolled in the study. Patients were grouped according to stage of chronic HBV infection as defined in the 2009 American Association for the Study of Liver Diseases (AASLD) and update of AASLD Practice Guidelines for Management of chronic hepatitis B (CHB), the EASL Clinical Practice Guidelines: Management of Chronic Hepatitis B and Guidelines on Prevention and Treatment of Chronic Hepatitis B in China 2005 (http://www.ncbi.nlm.nih.gov/pubmed/18167196). The five groups included: 1) inactive carriers (IC), 2) those with CHB, 3) cirrhosis, 4) HCC, and 5) uninfected healthy controls.
Inactive carriers were defined as individuals who tested positive for HBsAg and HBV-DNA with normal alanine aminotransferase (ALT) values on a minimum of three occasions over a one-year period. Only patients with HBV DNA levels of ≥50 IU/mL were selected to take part in the study
The chronic hepatitis group comprised individuals who tested positive for HBsAg, HBV-DNA, and either HBeAg or anti-HBe with persistently or intermittently elevated ALT values and histologic evidence of active necroinflammatory disease with fibrosis but not cirrhosis.
Cirrhosis was diagnosed on the basis of liver histology or typical radiologic features upon abdominal imaging. All cirrhotic patients were compensated, Child-Pugh A class.
The diagnosis of HCC was based on the revised version of diagnostic criteria for primary liver cancer in 1999.21 These criteria included: 1) alfa-fetoprotein (AFP)≥400 µg/L in the absence of pregnancy, reproductive or embryonic tumors, and a new space occupying lesion (SOL) within the liver having the characteristic findings of increased uptake of contrast during the arterial phase and early washout during the venous phase; 2) AFP<400 µg/L, a SOL with the above radiologic features plus the presence of vascular invasion or metastases; and/or 3) positive diagnostic cytology/histology.
Subjects were infected with C type HBV, and none of the chronic HBV-infected and cirrhotic patients had received prior antiviral treatment. Subjects were excluded if they had evidence of concurrent HCV, hepatitis D virus (HDV) or human immunodeficiency virus (HIV) infections, or alcohol-, and drug-induced, autoimmune or metabolic liver disorders, were receiving immunosuppressive therapy, or had significant associated co-morbidities (e.g., heart or kidney failure).
The study protocol was approved by the hospital ethics committee of the First Hospital of Jilin University.
Human genomic DNA was extracted from peripheral blood mononuclear cells (PBMCs) with a genomic DNA purification kit (Promega Co., Madison, WI, USA). Genotyping of the rs12979860, rs12980275, rs8103142, and rs8099917 polymorphisms was performed using a pyrosequencing method according to the manufacturer's protocol (PyroMark ID Pyrosequencing machine, Qiagen, Shanghai, China). The primers used for PCR were rs12979860: forward 5'-ATTC CTGGACGTGGATGGG TAC-3', reverse 5'-biotin-AGC GCGGAGTGCAATTCA-3'; rs8099917: forward 5'-TTGT CACTGTTCCTCCTTTTGTTT-3', reverse 5'-biotin-TGGGAGAATGCAAATGAGAGATA-3'; rs8103142: forward 5'-Biotin-AGAAGGTGAAGGGGCCACTA-3', reverse 5'-CTGTGCCTTTGCTGTCTAGGA-3'; rs12980275: forward 5'-GCCAGTCTCAAAAGAACAAATGC-3', reverse 5'-biotin-CTACCCCGGCAAATATTTAGACA-3'. Sequence primers for pyrosequencing analysis were rs 12979860: 5'-AGCTCCCCGAAGGCG-3'; rs8099917: 5'-TCCTTTCTGTGAGCAAT-3'; rs8103142: 5'-TGCTGA AGGACTGCA-3'; rs12980275: 5'-TGAGAGAAGTCA AATTCCT-3'.
PBMCs from 99 of the 156 subjects (26 CHB, 22 ICs, 17 cirrhosis, 17 HCC, and 17 healthy controls) were isolated using Lymphocyte Separating Solution (DingGuo Biotechnology Co., Shanghai, China). The remaining 57 subjects did not consent to provide additional venous blood. Total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA, USA), and cDNA was synthesized using random hexamers and SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions. Quantitative RT-PCR was performed using SYBR green (Realtime PCR MasterMix QPK-201, Toyobo, Osaka, Japan) on a 7300 Real-Time PCR System (Applied Biosystems, Grand Island, NY, USA). The expression levels of IL28A/B mRNA were normalized to median expression of GAPDH. Primers used were: IL28A/B: forward 5'-CTG CCA CAT AGC CCA GTT CAA GT-3', reverse 5'-ACT CTT CTA AGG CAT CTT TGG CCC-3', GAPDH: forward 5'-GAA GGT GAA GGT CGG AGTC-3', reverse 5'-GAA GAT GGT GAT GGG ATT TC-3'. The individual who performed gene expression analysis was blinded to the rs12979860 genotype being measured. To ensure that the amount of mRNA expression was not affected by other factors, RNA was immediately extracted upon isolation of PBMCs from blood and subjected to reverse transcription of cDNA and subsequent RT-PCR.
Serum IL28B levels were measured with an ELISA Kit specific for IL28B (Human λ3 IFN ELISA Kit, Catalogue number: I153-00, GBD Ltd., USA). The ELISA was carried out according to the manufacturer's instructions, and absorbance at 450 nm was measured with an ELISA-reader (Eppendorf® BioPhotometer, Hamburg, Germany). The sensitivity of the IL28B ELISA was 1 pg/mL and dynamic range; 1-1000 pg/mL. The intra-assay coefficient of variation was 3.7%, whereas the inter-assay variation was 5.2%.
Continuous variables were presented as mean (standard deviation) or median (interquartile), while categorical variables were expressed as frequencies (%). Differences between continuous variables were evaluated by the Student's t-test or ANOVA. Differences between categorical variables were assessed using the Pearson chi-square test, and continuous data using the Kruskal-Wallis nonparametric test. The chi-square G test "Goodness of Fit" was employed to verify whether the proportions of the polymorphism were distributed in the controls and HBV infection subjects in accordance with the Hardy-Weinberg equation. Differences in allelic and genotypic frequencies between groups were assessed by the chi-square test. Pairwise linkage disequilibrium and r2 between single nucleotide polymorphisms (SNPs) were calculated by PLINK software. A p-value<0.05 was considered significant (two-tailed). Statistical analysis of data was performed using the SPSS software package 18.0 (IBM, Armonk, NY, USA).
One hundred fifty-six Chinese subjects were enrolled in the study, including 50 IC, 34 CHB, 27 cirrhosis, 26 HCC, and 19 healthy controls. The frequencies of the key gene polymorphisms of IL28B (rs12979860, rs8099917, rs8103142, and rs12980275) were investigated in all 156 subjects. The linkage disequilibrium (LD) and haplotype analysis of the four SNPs were performed. The results showed that these SNPs were in complete LD (D'=0.99-1; r2=0.97-0.98); therefore, the rs12979860 polymorphism was selected as a tag SNP for the subsequent studies. The demographics, liver enzyme levels, genotype variants and viral loads are provided in Table 1. There were no significant differences in gender distributions but, as would be predicted, mean ages increased with more advanced stages of disease. Serum ALT values varied according to the stages of infection. In all stages, the C/C genotype was present at higher frequency than the C/T genotype. Viral loads were significantly higher in the CHB cohort versus other cohorts.
The results of PBMC IL28A/B mRNA expression at different disease stages are shown in Fig. 1. IL28A/B mRNA expression was higher in IC and healthy controls than CHB, cirrhosis, and HCC patients (IC vs. CHB, p=0.03; IC vs. cirrhosis, p<0.01; IC vs. HCC, p<0.01; controls vs. CHB, p=0.02; controls vs. cirrhosis, p<0.01; controls vs. HCC, p<0.01).
In addition, IL28A/B mRNA expression as a function of age, gender, viral load, and HBeAg status within the CHB cohort were analyzed. In this case, there was no association between IL28A/B mRNA expression and age, gender, viral load, or HBeAg status (data not shown).
Serum IL28B protein levels are provided in Fig. 2. No significant differences existed between the IC and control groups. However, the levels were significantly lower in CHB, cirrhosis, and HCC patients (IC vs. CHB, p=0.02; IC vs. cirrhosis, p=0.01; IC vs. HCC, p=0.001; controls vs. CHB, p<0.01; controls vs. IC, p<0.01; controls vs. HCC, p<0.01). When CHB patients were stratified according to HBeAg status, IL28B protein levels were higher in HBeAg-positive than negative patients (p=0.01) (Fig. 3). There were no associations between IL28B protein levels and age, gender, or viral loads (Table 2).
The rs12979860 variant upstream of the IL28B gene may affect IL28B protein production. Thus, we compared IL28B serum protein levels between C/C and T/C groups in all 156 subjects by ELISA. IL28B protein levels were significantly higher in the C/C group than the T/C group (C/C vs. T/C, p=0.005) (Fig. 4A). The same was true when only CHB patients were stratified with respect to rs12979860 genotypes (p=0.03) (Fig. 4B). Fig. 4 also provides the results of IL28A/B mRNA expression in PBMCs. Here again, IL28A/B mRNA expression was higher in C/C than T/C subjects (p=0.002) (Fig. 4C) as well as C/C and T/C subjects in the CHB and IC cohorts (p=0.01 and 0.019, respectively) (Fig. 4D and E). However, this was not the case in the cirrhosis, HCC, or control cohorts (data not shown).
Fig. 5 provides the results of IL28A/B mRNA expression (Fig. 5A) and serum IL28B protein levels (Fig. 5B) in the context of ALT values in the combined cohorts of 87 CHB, cirrhosis, and HCC patients. Both IL28A/B mRNA expression and IL28B protein levels were significantly higher in individuals with high (≥80 IU/mL) than low (<80 IU/mL) ALT values (p=0.04 and 0.001, respectively).
Table 2 describes the factors associated with high serum IL28B protein levels in HBV-infected patients as determined by multivariate logistic regression analysis. High IL28B levels were independently associated with the C/C genotype, individuals with high ALT levels, and mild liver disease.
The chromosomal region (19q13) encodes three cytokine genes (IL28A, IL28B, and IL29) that belong to the IFN-λ (also named type III IFN) family. IFN-λs interact with transmembrane receptors to induce potent antiviral responses that are mediated through activation of the JAK-STAT and MAPK pathways.22,23,24
In vitro and in vivo models have demonstrated the importance of IFN-λs in the immune response and up-regulation of transcription of IFN-stimulated genes required to control viral infections, including herpes simplex virus,25,26 HIV,27 and hepatitis B and C viruses.28,29 Of note, IFN-λs have also been reported to inhibit HBV replication in an experimental model.29
The rs12979860 allele is 3 kb upstream from the IL28B locus, which encodes several genes including IL28A and IL29.22,23 Thus, it is likely that the SNP may also affect the function of other genes in the locus. Indeed, it has been reported that this variant is associated with increased serum IL29 and IL28A/B levels and resolution of HCV infections.30 These findings suggest that variations of upstream of the IL28B gene may impact the expression and production of all IFN-λs, which may explain, in part, their association with different outcomes of HBV infection. It is important to further elucidate the mechanism by which gene variants regulate the expression of IFN-λs in HBV infection.
Although the influences of SNPs on IL28B gene expression and antiviral activity have been extensively studied in patients with HCV infections,31 relatively little is known about the role gene variants played in IL28B production and outcomes in patients with HBV infections. Recently, Lee, et al.32 reported that the IL28B rs10853728 C/C genotype is associated with active hepatitis in HBeAg-negative CHB, and that host factors play a role in disease activity during the different phases of CHB. Results from our present study indicate that IL28A/B mRNA expression and IL28B protein levels are significantly lower in patients with active or advanced disease (CHB, cirrhosis, and HCC) when compared to those with inactive disease (IC) or healthy controls. These findings were supported by the observation of greater IL28A/B mRNA expression and higher serum IL28B protein levels in HBeAg-positive than -negative patients (HBeAg positivity is most common in patients with early, inactive liver disease). We also confirmed a previous study by Ren, et al.33 that HBV-infected individuals with the rs12979860 C/C genotype have higher IL28A/B mRNA and IL28B protein levels than those carrying the T-alleles. Together, these results suggest that patients with reduced IL28B expression tend to have more active and advanced stage liver disease, and that IL28B variants have an effect on IL28B production.
Serum ALT levels are often used to monitor necroinflammatory disease activity in patients with chronic HBV infections. We found that IL28A/B mRNA expression was significantly higher in those patients with active or advanced stages of the disease (CHB, cirrhosis, and HCC) and high serum ALT levels. The significance of this observation and the precise relationship between IL28B and ALT levels in HBV infection is unknown. Since IL28B is involved in antiviral immunity, it is tempting to speculate that higher serum ALT and IL28B levels reflect enhanced virus-host interactions. If so, the combination of ALT and IL28B levels could serve as an additional predicator of the outcome of chronic HBV infections.
When incorporated into logistic regression analysis, the factors most associated with high serum IL28B protein levels were the C/C genotype, high ALT levels, and inactive disease. After adjusting for ALT levels and stage of disease in the multivariate logistic regression analysis, the rs12979860 C/C genotype remained independently associated with high IL28B protein levels.
Additional prospective studies are required to determine whether low IL28A/B mRNA expression and IL28 protein levels in patients with active or advanced disease (the CHB, cirrhotic, and HCC cohorts) reflect the cause or effect of the disease stage. The findings that the C/C genotype was equally distributed across all patient cohorts, and that viral loads were not associated with IL28B protein levels suggest that chronic necroinflammatory disease and/or hepatic dysfunction attenuate IL28A/B mRNA expression, thereby resulting in lower IL28B protein levels (despite the C/C genotype profile). If so, this could reflect a successful attempt by the virus to evade IFN-λ-mediated immune clearance.
In conclusion, our results indicate that IL28A/B mRNA expression and IL28B protein levels may correlate with the activity and long-term stage of chronic HBV infections in Chinese Han patients. Furthermore, the SNP rs12979860 upstream of IL28B is likely associated with enhanced IL28B production. Additional research is required to reveal any cause-and-effect relationship between these polymorphisms and host protective immunity against HBV, and whether HBV infection or liver disease results in down-regulated IL28B expression. According to the association between the presence of the IL28B allele and response to interferon and ribavirin treatment with HCV infection,34 further research is required to demonstrate that IL28B genotype may be associated with treatment response in HBV patients.
ACKNOWLEDGEMENTS
We would like to thank the nursing staff at the First Hospital of Jilin University for making this study possible, and Mrs. Helen Arthur for critical reading of the manuscript.
This study was supported by the National Natural Science Foundation of China (grants No. 81001312/H1005 and 81072347/H2609), the National Natural Science Foundation of China (grant No. 30972611), and the National Science and Technology Major Project 2012ZX10002003.
References
1. Margolis HS. Hepatitis B virus infection. Bull World Health Organ. 1998; 76(Suppl 2):152–153. PMID: 10063701.
2. Chen CJ, Wang LY, Yu MW. Epidemiology of hepatitis B virus infection in the Asia-Pacific region. J Gastroenterol Hepatol. 2000; 15:E3–E6. PMID: 10921373.

3. Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology. 2009; 50:661–662. PMID: 19714720.

4. El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007; 132:2557–2576. PMID: 17570226.

5. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005; 55:74–108. PMID: 15761078.

6. Yim HJ, Lok AS. Natural history of chronic hepatitis B virus infection: what we knew in 1981 and what we know in 2005. Hepatology. 2006; 43(2 Suppl 1):S173–S181. PMID: 16447285.

7. Thio CL, Thomas DL, Carrington M. Chronic viral hepatitis and the human genome. Hepatology. 2000; 31:819–827. PMID: 10733534.

8. Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet. 2009; 41:591–595. PMID: 19349983.

9. Guo Y, Guo H, Zhang L, Xie H, Zhao X, Wang F, et al. Genomic analysis of anti-hepatitis B virus (HBV) activity by small interfering RNA and lamivudine in stable HBV-producing cells. J Virol. 2005; 79:14392–14403. PMID: 16254373.

10. Mbarek H, Ochi H, Urabe Y, Kumar V, Kubo M, Hosono N, et al. A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum Mol Genet. 2011; 20:3884–3892. PMID: 21750111.

11. Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009; 41:1100–1104. PMID: 19749758.
12. Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009; 461:399–401. PMID: 19684573.

13. Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009; 461:798–801. PMID: 19759533.

14. Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009; 41:1105–1109. PMID: 19749757.
15. Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, et al. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci U S A. 2008; 105:7034–7039. PMID: 18467494.

16. Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD, et al. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010; 52:1888–1896. PMID: 20931559.

17. Sonneveld MJ, Wong VW, Woltman AM, Wong GL, Cakaloglu Y, Zeuzem S, et al. Polymorphisms near IL28B and serologic response to peginterferon in HBeAg-positive patients with chronic hepatitis B. Gastroenterology. 2012; 142:513–520. PMID: 22108195.

18. Peng LJ, Guo JS, Zhang Z, Shi H, Wang J, Wang JY. IL28B rs12979860 polymorphism does not influence outcomes of hepatitis B virus infection. Tissue Antigens. 2012; 79:302–305. PMID: 22239156.

19. Li W, Jiang Y, Jin Q, Shi X, Jin J, Gao Y, et al. Expression and gene polymorphisms of interleukin 28B and hepatitis B virus infection in a Chinese Han population. Liver Int. 2011; 31:1118–1126. PMID: 21745278.

20. Noureddin M, Wright EC, Alter HJ, Clark S, Thomas E, Chen R, et al. Association of IL28B genotype with fibrosis progression and clinical outcomes in patients with chronic hepatitis C: a longitudinal analysis. Hepatology. 2013; 58:1548–1557. PMID: 23703931.

21. Poon D, Anderson BO, Chen LT, Tanaka K, Lau WY, Van Cutsem E, et al. Management of hepatocellular carcinoma in Asia: consensus statement from the Asian Oncology Summit 2009. Lancet Oncol. 2009; 10:1111–1118. PMID: 19880065.

22. Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003; 4:63–68. PMID: 12469119.

23. Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003; 4:69–77. PMID: 12483210.
24. Li M, Liu X, Zhou Y, Su SB. Interferon-lambdas: the modulators of antivirus, antitumor, and immune responses. J Leukoc Biol. 2009; 86:23–32. PMID: 19304895.
25. Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006; 80:4501–4509. PMID: 16611910.
26. Melchjorsen J, Sirén J, Julkunen I, Paludan SR, Matikainen S. Induction of cytokine expression by herpes simplex virus in human monocyte-derived macrophages and dendritic cells is dependent on virus replication and is counteracted by ICP27 targeting NF-kappaB and IRF-3. J Gen Virol. 2006; 87(Pt 5):1099–1108. PMID: 16603509.
27. Hou W, Wang X, Ye L, Zhou L, Yang ZQ, Riedel E, et al. Lambda interferon inhibits human immunodeficiency virus type 1 infection of macrophages. J Virol. 2009; 83:3834–3842. PMID: 19193806.

28. Hong SH, Cho O, Kim K, Shin HJ, Kotenko SV, Park S. Effect of interferon-lambda on replication of hepatitis B virus in human hepatoma cells. Virus Res. 2007; 126:245–249. PMID: 17451832.

29. Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005; 79:3851–3854. PMID: 15731279.

30. Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010; 138:1338–1345. PMID: 20060832.

31. Langhans B, Kupfer B, Braunschweiger I, Arndt S, Schulte W, Nischalke HD, et al. Interferon-lambda serum levels in hepatitis C. J Hepatol. 2011; 54:859–865. PMID: 21145813.

32. Lee IC, Lin CH, Huang YH, Huo TI, Su CW, Hou MC, et al. IL28B polymorphism correlates with active hepatitis in patients with HBeAg-negative chronic hepatitis B. PLoS One. 2013; 8:e58071. PMID: 23469142.

33. Ren S, Lu J, Du X, Huang Y, Ma L, Huo H, et al. Genetic variation in IL28B is associated with the development of hepatitis B-related hepatocellular carcinoma. Cancer Immunol Immunother. 2012; 61:1433–1439. PMID: 22310928.
34. Shi X, Pan Y, Wang M, Wang D, Li W, Jiang T, et al. IL28B genetic variation is associated with spontaneous clearance of hepatitis C virus, treatment response, serum IL-28B levels in Chinese population. PLoS One. 2012; 7:e37054. PMID: 22649509.

Fig. 1
The association of IL28A/B mRNA expression with different outcomes of chronic HBV. IL28A/B mRNA expression was documented in peripheral blood mononuclear cells (PBMCs) from patients with CHB (n=26), IC (n=22), cirrhosis (n=17), HCC (n=17), and healthy controls (n=17). Data provided represent the median (quartile range) and are representative of two determinations. Some subjects did not consent to provide additional venous blood, therefore, PBMCs from 99 of the 156 subjects were isolated. IC, inactive carriers; CHB, chronic hepatitis B; LC, liver cirrhosis; IL, interleukin.
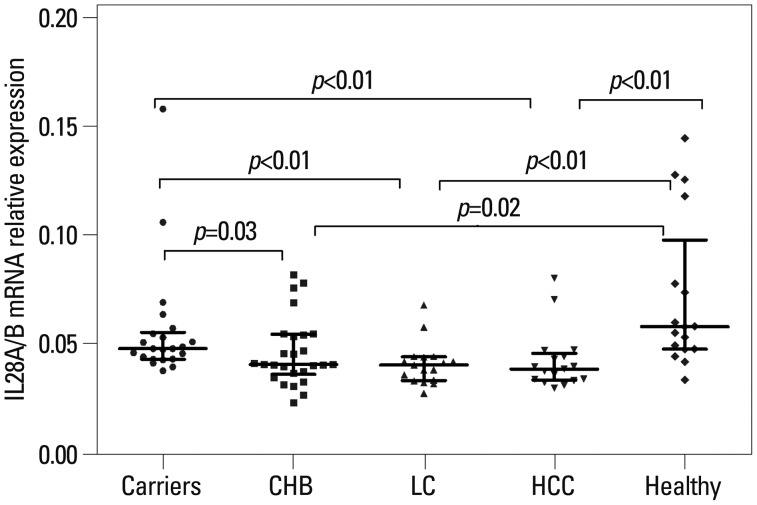
Fig. 2
The association of serum IL28B protein levels with different outcomes of chronic HBV. Data provided represent the median (quartile range) and are representative of two determinations. IC, inactive carriers; CHB, chronic hepatitis B; HCC, hepatocellular carcinoma; IL, interleukin.
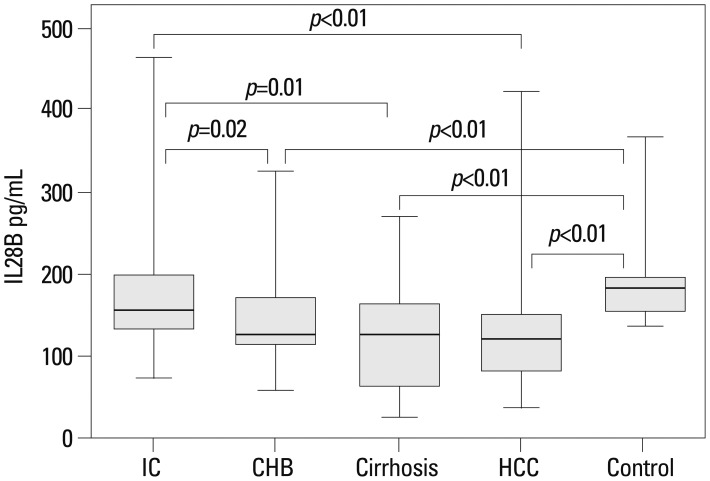
Fig. 3
Serum IL28B protein levels in 34 patients with chronic hepatitis B (CHB) and HBeAg positive (e+) or negative (e-) serology. HBeAg positive (n=22) and HBeAg negative (n=12). Data provided represent the median (quartile range) and are representative of two determinations. IL, interleukin.
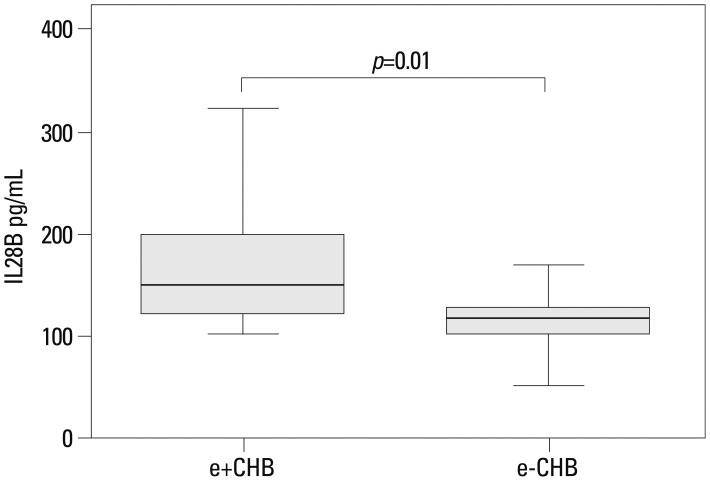
Fig. 4
The association of IL28A/B genotype and serum IL28B protein levels relative to C/C and T/C genotypes mRNA expression of IL28A/B. (A) IL28B serum protein levels in all 156 subjects. (B) IL28B serum protein levels in the CHB cohort. (C) IL28A/B mRNA expression in all 99 subjects by qRT-PCR. (D and E) IL28A/B mRNA in the CHB and IC cohorts, respectively. Data provided represent the median (quartile range) and are representative of two determinations. IC, inactive carriers; CHB, chronic hepatitis B.
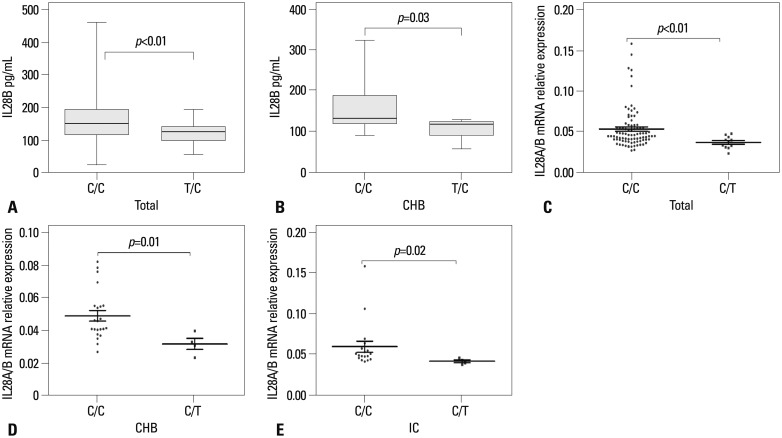
Fig. 5
(A) The association between IL28A/B mRNA expression (high ALT patients 34, low ALT patients 26) in patients with advanced stage disease and high versus low serum ALT levels. (B) The association between IL28B serum protein levels (high ALT patients 45, low ALT patients 42) in patients with advanced stage disease and high versus low serum ALT levels. Data provided represent the median (quartile range) and are representative of two determinations. ALT, alanine aminotransferase; CHB, chronic hepatitis B; HCC, hepatocellular carcinoma.
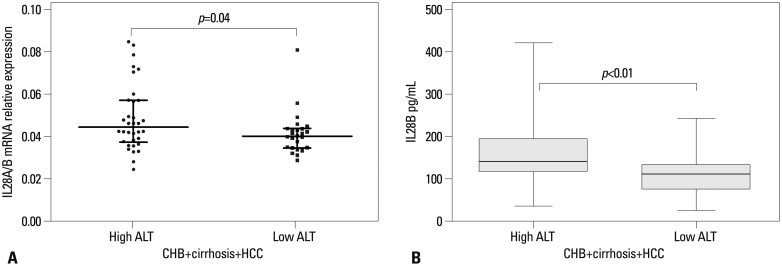
Table 1
Baseline Characteristic of 156 Study Subjects

SD, standard deviation; CHB, chronic hepatitis B; HCC, hepatocellular carcinoma; HBV, hepatitis B virus; ALT, alanine aminotransferase.
IC: Chronic HBV carrier: HBsAg and HBeAg positive, higher HBV DNA and normal ALT; Inactive HBsAg carrier: HBeAg negative, anti-HBe positive, low or undetectable HBV DNA and normal ALT.
*Median (quartile).
†Comparison across IC, CHB, cirrhosis, and HCC.
‡Comprison among CHB, cirrhosis, and HCC.
Table 2
Factors Associated with High IL28B Serum Level in HBV Infected Patients by Multivariate Analysis





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download