Abstract
Purpose
Although the polymorphisms of erythrocyte complement receptor type 1 (CR1) in patients with malaria have been extensively studied, a question of whether the polymorphisms of CR1 are associated with severe malaria remains controversial. Furthermore, no study has examined the association of CR1 polymorphisms with malaria in Chinese population. Therefore, we investigated the relationship of CR1 gene polymorphism and malaria in Chinese population.
Materials and Methods
We analyzed polymorphisms of CR1 gene rs2274567 G/A, rs4844600 G/A, and rs2296160 C/T in 509 patients with malaria and 503 controls, using the Taqman genotyping assay and PCR-direct sequencing.
Results
There were no significant differences in the genotype, allele and haplotype frequencies of CR1 gene rs2274567 G/A, rs4844600 G/A, and rs2296160 C/T polymorphisms between patients with malaria and controls. Furthermore, there was no association of polymorphisms in the CR1 gene with the severity of malaria in Chinese population.
The global malaria is still very serious in currently. From the World Health Organization (WHO) report in 2008 indicated that an estimated 247 million malaria cases and about 3.3 billion people at risk.1 The manifestations of malaria range from asymptomatic parasitaemia in semi-immune individuals to severe disease in susceptible populations, including the children under the age of five years, pregnant women, and non-immune adults. Among the infected patients, the symptoms of severe malaria have shown on different forms, including severe anaemia, cerebral malaria, placental malaria, metabolic acidosis, and hypoglycaemia, and so on.2,3 Although there were variety of causes to cause those disease patterns, but recent researches have shown that lack of erythrocyte complement regulatory proteins was the main in pathogenesis of severe malaria anaemia, those complement regulatory proteins including complement receptor 1 and CD55 (CR1/CD35).4,5 Those complement regulatory proteins can inhibit formation of C3 convertases, and prevent terminal polymerization of membrane attack complex, thus protecting the destruction of red blood cells.6
CR1 is an immune-regulatory protein found on the surface of erythrocytes and most leukocytes, and its functions include key complement regulation of complement activation and clearance of immune complexes.7,8 Furthermore, increasing evidences from several studies suggest that CR1 plays a critical role in the pathogenesis of Plasmodium falciparum (P. falciparum) malaria.9,10 Early study found that the surface of erythrocytes of CR1 can bind the infected red cells to uninfected red cells to form rosettes, which is thought to contribute to malaria pathology by causing microvascular obstruction and impaired tissue perfusion.11 Recently, studies indicated also that CR1 is a receptor for the invasion of erythrocytes by P. falciparum,12 and some studies suggest that CR1 not only regulate the complement system but also clear the immune complexes during malaria infection.13,14 Thus, the correlation of CR1 receptor and malaria remains to be studied.11
The gene encoding CR1 is located at long arm of chromosome 1 in humans. Recent data showed that CR1 gene polymorphisms influence CR1 production or protein expression and are associated with malaria in several populations.15,16,17 Furthermore, clinical studies showed a correlation between the levels of serum CR1 and disease severity in patients with malaria.18,19 On the other hand, some results were inconsistent.20,21 Moreover, the CR1 expression level in the populations from Papua New Guinea (PNG), Mali, and the United Kingdom has recently been shown to be not associated with polymorphisms in the 3' untranslated regions of the CR1 gene.22 Although, little is known about the role of single nucleotide polymorphisms (SNPs) of CR1 gene in the development of malaria, National Center for Biotechnology Information SNP database showed that CR1 gene rs2274567 G/A, rs4844600 G/A, and rs2296160 C/T sites (all are located on coding region) have frequencies of polymorphism distribution in different ethnicity, including Chinese. Lastly, research for Thailand population indicated that the CR1 gene rs2274567 G/A and rs2296160 C/T sites polymorphisms are not associated with CR1 level and malaria.23 However, there have been no studies to examine the association between SNPs of CR1 gene rs2274567 G/A, rs4844600 G/A, and rs2296160 C/T with malaria in a Chinese population. In this study, we evaluated whether CR1 gene rs2274567 G/A, rs4844600 G/A, rs2296160 C/T sites polymorphisms and haplotype are associated with malaria in a Chinese population.
The case-control population contained 1012 adult unrelated Chinese selected from the same populations living in China between February and August 2012, and the demographics of the cases and controls are shown in Table 1. There were no significant differences between the cases and controls for the mean age or gender distribution, smoking and alcohol consumption status, suggesting that the matching based on these two variables was adequate. For all patients with malaria, malarial infection with P. falciparum was confirmed by a positive blood smear result for the asexual form of P. falciparum. Clinical manifestations of malaria were classified according to the definitions and associated criteria set forth by the WHO. The patients were selected from groups with 3 different types of malaria (i.e., mild malaria, noncerebral severe malaria, and cerebral malaria). The mild malaria group consisted of 327 patients with mild symptoms who were characterized by fever without other causes of infection but by none of the manifestations of severe malaria described below. The noncerebral severe malaria group (hereafter referred to the "severe malaria group") comprised 102 patients with severe malaria, which was defined as malaria with evidence of vital organ dysfunction or with one of the following signs of severity: a high level of parasitemia (>100000 parasites/mL), hypoglycemia (plasma glucose level <22 nmol/L), severe anemia (a hematocrit of <20% or a hemoglobin level <7 g/dL), or a serum creatinine level >3 mg/dL. The cerebral malaria group consisted of 80 patients who experienced a coma from which they could not be roused and for whom noncerebral causes of coma could be excluded. All of the patients in this group were ≥9 years of age, and mean ages of patients with mild malaria, severe malaria, and cerebral malaria were 43.1±12.3, 40.2±9.8, and 43.7±14.2 years, respectively. The control group comprised 503 healthy volunteers selected on the basis of no evidence of any personal or family history of malaria or other serious illness. The mean age of the control group (247 males and 256 females) was 43.1±13.5 years. Written informed consent was obtained from all the subjects, and the study was performed with the approval of the ethics committee of Chinese Human Genome.
Genomic DNA was extracted from ethylenediaminetetraacetic acid-anticoagulated peripheral blood leukocytes by the salting-out method.24 Briefly, 3 mL of blood was mixed with Triton lysis buffer (0.32 M sucrose, 1% Triton X 100, 5 mM MgCl2, H2O, 10 mM Tris-HCl, pH 7.5). Leucocytes were spun down and washed with H2O. The pellet was incubated with proteinase K at 56℃ and subsequently salted out at 4℃ using a saturated NaCl solution. Precipitated proteins were removed by centrifugation. The DNA in the supernatant fluid was dissolved in 300 µL of H2O.
The oligonucleotide forward primer 5'-AGC AAA ACT TTC ATA AAA CTT ACC AAC-3' and reverse primer 5'-CGC AGA AGG TAA AAT GAG AGT ATG T-3' were used to amplify a fragment, which included CR1 gene rs2296160 site, by polymerase chain reaction (PCR). Similarly, using PCR, forward primer 5'-GCT CCC TGA CAG TCC CTC CT-3' and revere primer 5'-CAG CAT AGA CAT AGA GCA CGA GG-3' were used for a fragment which included CR1 gene rs2274567 site, and forward primer 5'-AGC AAA ACT TTC ATA AAA CTT ACC AAC-3' and revere primer 5'-CGC AGA AGG TAA AAT GAG AGT ATG T-3' were used for a fragment which included CR1 gene rs4844600. All three sites of CR1 gene production by PCR were used for DNA sequencing. The genotyping was performed on an ABI 7900 real-time PCR by Taqman allele discrimination method (Taqman assay; Applied Biosystems, Shanghai, China) using the manufacturers specifications. The Taqman genotyping assay was supplied by Applied Biosystems. The genotypings were confirmed by PCR-direct sequencing using sequenator of ABI 3500 (DNA sequencing; Applied Biosystems, Shanghai, China).
Genotype and allele frequencies of CR1 were compared between malaria cases and controls using the χ2 test and Fisher's exact test when appropriate, and odds ratios and 95% confidence intervals were calculated to assess the relative risk conferred by a particular allele and genotype. Demographic and clinical data between groups were compared by χ2 test and by Student's t-test. Hardy-Weinberg equilibrium was tested for with a goodness of fit χ2-test with one degree of freedom to compare the observed genotype frequencies among the subjects with the expected genotype frequencies. The linkage disequilibrium (LD) between the polymorphisms was quantified using the Shi's standardized coefficient D' (|D'|).25 The haplotypes and their frequencies were estimated based on a Bayesian algorithm using the Phase program.26 Statistical significance was assumed at the p<0.05 level. The SPSS statistical software package version 11.5 (SPSS Inc., Chicago, IL, USA) was used for all of the statistical analysis.
The results of sequencing, including the CR1 gene rs2274567 G/A, rs4844600 G/A, and rs2296160 C/T sites, are shown in Fig. 1. The genotype and allele frequencies of the CR1 gene rs2274567 G/A, rs4844600 G/A, and rs2296160 C/T polymorphisms in the group of patients with malaria and in the control group are shown in Table 2. The genotype distributions of three polymorphisms among the controls and the cases were in Hardy-Weinberg equilibrium. There were no significant differences in the genotype and allele frequencies of the CR1 gene rs2274567 G/A, rs4844600 G/A, and rs2296160 C/T polymorphisms between the two study groups (p>0.05).
No association was found between CR1 gene rs2274567 G/A, rs4844600 G/A, and rs2296160 C/T polymorphisms and different types of malaria, shown in Table 3. Genotype and allele frequencies of the CR1 gene rs2274567 G/A, rs4844600 G/A, and rs2296160 C/T polymorphisms in mild malaria were not significantly different between cerebral malaria & severe malaria of malaria patients, respectively (p>0.05).
Haplotype analyses were performed, and the possible eight haplotype frequencies are shown in Table 4. The rs2274567 G/A polymorphism was in strong LD with the rs4844600 G/A (|D'|=0.865) and rs2296160 C/T (|D'|=0.854), respectively. The rs4844600 G/A and rs2296160 C/T were in strong LD (|D'|=0.871). Major GGC haplotype accounted for 50.2% and 49.4% of these eight haplotypes in both patients and controls, respectively. However, haplotype frequencies of the CR1 gene in malaria patients were not significantly different than that in healthy controls (p>0.05).
To our best knowledge, this is the first attempts to evaluate the association between the single-nucleotide polymorphism and haplotype of CR1 gene rs2274567 G/A, rs4844600 G/A, and rs2296160 C/T sites and malaria in a Chinese population, and we failed to observe an association of CR1 genotypes, alleles and haplotypes with malaria or any subtype of malaria in Chinese population. These data suggest that CR1 gene rs2274567 G/A, rs4844600 G/A, and rs2296160 C/T sites polymorphisms may not be an important contributor to malaria in Chinese population.
As a Complete membrane protein, CR1 mainly found on red blood cells, lymphocytes, macrophages, follicular dendritic cells, macrophages, and kidney podocytes, and so on. Some of functions were found on CR1, including to bind to and remove the immune complexes of C4b- and C3b-, and as a complement regulatory protein 1. Recent research indicated that the CR1 could connect the uninfected red blood cells to resetting of P. falciparum-infected red blood cells through banding to P. falciparum erythrocyte membrane protein 1.27 Several studies have shown that in vitro rosette formation was closely related to the severity of malaria, and CR1 gene polymorphism may be to play an important role on susceptibility to severe malaria pathogenesis.28,29 Furthermore, some data have demonstrated that erythrocytes can play a dual role in immune regulation, removing immune complexes. Single nucleotide polymorphisms in the promoter of CR1 gene would reduce the level of erythrocyte complement receptor type 1 (E-CR1) expression, leading to the accumulation of immune complexes. A lower E-CR1 expression causes tissue damage or cell aggregation and this would lead to severe clinical features, including cerebral malaria. On the other hand, it's higher expression of would protect the development of severe symptoms through improving the clearance rate of immune complexes. Nevertheless, several studies showed contradictory results; higher number of copies of E-CR1 increased the rosette formation,30,31,32,33 and increased rosette formation was associated with severe clinical syndrome of malaria infection.34,35,36,37 Research indicated that the genetic polymorphisms in exon 19 (3093G>T), exon 22 (3650A>G), exon 33 (5507C>G), and intron 27 (Hind III A>T) of the CR1 gene were associated with the level of CR1 expression in different populations of malaria patients.38 And study also shown that contains A Hind III, G3093, A3650, and C5507 of haplotype allele (high expression haplotype) could increase the level expression of CR1, and contains T Hind III, T3093, G3650, and G5507 of haplotype allele (low expression haplotype) would decrease the level expression of CR1.38 Population survey of malaria-endemic area indicated that have some conflicting findings on the association of CR1 polymorphism with malaria, on the one hand, with the intermediate CR1 level of heterozygotes (HL) individuals for the CR1 exon 22 polymorphism significantly protected against severe malaria in PNG,15 on the other hand, with the low CR1 level of homozygous (LL) individuals for the CR1 intron 27 were significantly higher in severe malaria.16 Furthermore, study also shown that the CR1 of rs9429942-T was associated with a high level of expression of erythrocyte-CR1 and contributes to protect against cerebral malaria in Thailand.23 However, the intron 27 and exon 33 variants of CR1 gene did not influence the level of erythrocyte-CR1 expression in African population.21 In the present study, we found no apparent relationship of the CR1 gene CR1 gene rs2274567 G/A, rs4844600 G/A, and rs2296160 C/T sites polymorphisms with the risk of malaria. No strict group for server malaria, thus result in underestimate or overestimate the actual level of CR1 might be the reason of the difference of clinical symptoms and CR1 genotype in different studies. Researches indicated that age-related and/or regional differences of malaria endemic area would change CR1 level on RBC.39,40 In addition, non-random sampling, a limited sample size, and the possible selection bias might be influencing the outcome of different researches. In this study, we also cannot exclude the possibility that observed association depends on a gene in LD with the CR1 gene or effect of CR1 expression on other factors.
In summary, we did not find an association between CR1 gene rs2274567 G/A, rs4844600 G/A, and rs2296160 C/T sites polymorphisms and malaria or any subtype severity of malaria in a Chinese population. Nevertheless, further studies are needed to explore the complex interaction between environmental factors and CR1 gene polymorphisms in the risk of malaria, especially in ethnically disparate populations.
Figures and Tables
Fig. 1
PCR-direct sequencing. CR1 gene rs2274567 site genotype GG, GA, and AA (A). CR1 gene rs4844600 site genotype GG, GA, and AA (B). CR1 gene rs2296160 site genotype CC, CT, and TT (C). PCR, polymerase chain reaction; CR1, complement receptor type 1.

Table 1
Clinical Characteristics of the Study Participants

Table 2
The Genotype and Allele Frequencies of CR1 Polymorphisms in Malaria Patients and Controls
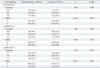
Table 3
The Genotype and Allele Frequencies of CR1 Polymorphisms in Relation to Malaria Severity
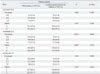
Table 4
Haplotype Frequencies of CR1 Gene in the Patients with Malaria and in Controls

ACKNOWLEDGEMENTS
This study was supported by National Natural Science Foundation (No. 81160212), the Key Project of Guangxi bureau of health, Guangxi, China (No. 200986), and the Open Fund in Medical Laboratory Science Center of Guangxi (kfjj 2011-03).
References
2. Schofield L, Grau GE. Immunological processes in malaria pathogenesis. Nat Rev Immunol. 2005; 5:722–735.

3. Mackintosh CL, Beeson JG, Marsh K. Clinical features and pathogenesis of severe malaria. Trends Parasitol. 2004; 20:597–603.

4. Stoute JA, Odindo AO, Owuor BO, Mibei EK, Opollo MO, Waitumbi JN. Loss of red blood cell-complement regulatory proteins and increased levels of circulating immune complexes are associated with severe malarial anemia. J Infect Dis. 2003; 187:522–525.

5. Waitumbi JN, Opollo MO, Muga RO, Misore AO, Stoute JA. Red cell surface changes and erythrophagocytosis in children with severe plasmodium falciparum anemia. Blood. 2000; 95:1481–1486.

6. Vorup-Jensen T, Boesen T. Protein ultrastructure and the nanoscience of complement activation. Adv Drug Deliv Rev. 2011; 63:1008–1019.

7. Lapin ZJ, Höppener C, Gelbard HA, Novotny L. Near-field quantification of complement receptor 1 (CR1/CD35) protein clustering in human erythrocytes. J Neuroimmune Pharmacol. 2012; 7:539–543.

8. Nuutila J, Jalava-Karvinen P, Hohenthal U, Kotilainen P, Pelliniemi TT, Nikoskelainen J, et al. Use of complement regulators, CD35, CD46, CD55, and CD59, on leukocytes as markers for diagnosis of viral and bacterial infections. Hum Immunol. 2013; 74:522–530.

9. Awandare GA, Spadafora C, Moch JK, Dutta S, Haynes JD, Stoute JA. Plasmodium falciparum field isolates use complement receptor 1 (CR1) as a receptor for invasion of erythrocytes. Mol Biochem Parasitol. 2011; 177:57–60.

11. Newbold C, Craig A, Kyes S, Rowe A, Fernandez-Reyes D, Fagan T. Cytoadherence, pathogenesis and the infected red cell surface in Plasmodium falciparum. Int J Parasitol. 1999; 29:927–937.

12. Spadafora C, Awandare GA, Kopydlowski KM, Czege J, Moch JK, Finberg RW, et al. Complement receptor 1 is a sialic acid-independent erythrocyte receptor of Plasmodium falciparum. PLoS Pathog. 2010; 6:e1000968.

13. Fernandez-Arias C, Lopez JP, Hernandez-Perez JN, Bautista-Ojeda MD, Branch O, Rodriguez A. Malaria inhibits surface expression of complement receptor 1 in monocytes/macrophages, causing decreased immune complex internalization. J Immunol. 2013; 190:3363–3372.

14. Thomas BN, Diallo DA, Noumsi GT, Moulds JM. Circulating Immune Complex Levels are Associated with Disease Severity and Seasonality in Children with Malaria from Mali. Biomark Insights. 2012; 7:81–86.

15. Cockburn IA, Mackinnon MJ, O'Donnell A, Allen SJ, Moulds JM, Baisor M, et al. A human complement receptor 1 polymorphism that reduces Plasmodium falciparum rosetting confers protection against severe malaria. Proc Natl Acad Sci U S A. 2004; 101:272–277.

16. Nagayasu E, Ito M, Akaki M, Nakano Y, Kimura M, Looareesuwan S, et al. CR1 density polymorphism on erythrocytes of falciparum malaria patients in Thailand. Am J Trop Med Hyg. 2001; 64:1–5.

17. Rout R, Dhangadamajhi G, Mohapatra BN, Kar SK, Ranjit M. High CR1 level and related polymorphic variants are associated with cerebral malaria in eastern-India. Infect Genet Evol. 2011; 11:139–144.

18. Mahajan RC, Narain K, Mahanta J. Anaemia & expression levels of CD35, CD55 & CD59 on red blood cells in Plasmodium falciparum malaria patients from India. Indian J Med Res. 2011; 133:662–664.
19. Tham WH, Schmidt CQ, Hauhart RE, Guariento M, Tetteh-Quarcoo PB, Lopaticki S, et al. Plasmodium falciparum uses a key functional site in complement receptor type-1 for invasion of human erythrocytes. Blood. 2011; 118:1923–1933.

20. Soares SC, Abé-Sandes K, Nascimento Filho VB, Nunes FM, Silva WA Jr. Genetic polymorphisms in TLR4, CR1 and Duffy genes are not associated with malaria resistance in patients from Baixo Amazonas region, Brazil. Genet Mol Res. 2008; 7:1011–1019.

21. Rowe JA, Raza A, Diallo DA, Baby M, Poudiougo B, Coulibaly D, et al. Erythrocyte CR1 expression level does not correlate with a HindIII restriction fragment length polymorphism in Africans; implications for studies on malaria susceptibility. Genes Immun. 2002; 3:497–500.

22. Cockburn IA, Rowe JA. Erythrocyte complement receptor 1 (CR1) expression level is not associated with polymorphisms in the promoter or 3' untranslated regions of the CR1 gene. Int J Immunogenet. 2006; 33:17–20.

23. Teeranaipong P, Ohashi J, Patarapotikul J, Kimura R, Nuchnoi P, Hananantachai H, et al. A functional single-nucleotide polymorphism in the CR1 promoter region contributes to protection against cerebral malaria. J Infect Dis. 2008; 198:1880–1891.

24. John SW, Weitzner G, Rozen R, Scriver CR. A rapid procedure for extracting genomic DNA from leukocytes. Nucleic Acids Res. 1991; 19:408.

25. Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005; 15:97–98.

26. Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001; 68:978–989.

27. Kirchgatter K, Portillo Hdel A. Association of severe noncerebral Plasmodium falciparum malaria in Brazil with expressed PfEMP1 DBL1 alpha sequences lacking cysteine residues. Mol Med. 2002; 8:16–23.

28. Valle-Delgado JJ, Urbán P, Fernàndez-Busquets X. Demonstration of specific binding of heparin to Plasmodium falciparum-infected vs. non-infected red blood cells by single-molecule force spectroscopy. Nanoscale. 2013; 5:3673–3680.

29. Doumbo OK, Thera MA, Koné AK, Raza A, Tempest LJ, Lyke KE, et al. High levels of Plasmodium falciparum rosetting in all clinical forms of severe malaria in African children. Am J Trop Med Hyg. 2009; 81:987–993.

30. Krych-Goldberg M, Moulds JM, Atkinson JP. Human complement receptor type 1 (CR1) binds to a major malarial adhesin. Trends Mol Med. 2002; 8:531–537.

31. Rowe JA, Rogerson SJ, Raza A, Moulds JM, Kazatchkine MD, Marsh K, et al. Mapping of the region of complement receptor (CR) 1 required for Plasmodium falciparum rosetting and demonstration of the importance of CR1 in rosetting in field isolates. J Immunol. 2000; 165:6341–6346.

32. Rowe JA, Moulds JM, Newbold CI, Miller LH. P. falciparum rosetting mediated by a parasite-variant erythrocyte membrane protein and complement-receptor 1. Nature. 1997; 388:292–295.

33. Stoute JA. Complement-regulatory proteins in severe malaria: too little or too much of a good thing? Trends Parasitol. 2005; 21:218–223.

34. Carlson J, Helmby H, Hill AV, Brewster D, Greenwood BM, Wahlgren M. Human cerebral malaria: association with erythrocyte rosetting and lack of anti-rosetting antibodies. Lancet. 1990; 336:1457–1460.

35. Treutiger CJ, Hedlund I, Helmby H, Carlson J, Jepson A, Twumasi P, et al. Rosette formation in Plasmodium falciparum isolates and anti-rosette activity of sera from Gambians with cerebral or uncomplicated malaria. Am J Trop Med Hyg. 1992; 46:503–510.

36. Heddini A, Pettersson F, Kai O, Shafi J, Obiero J, Chen Q, et al. Fresh isolates from children with severe Plasmodium falciparum malaria bind to multiple receptors. Infect Immun. 2001; 69:5849–5856.

37. Rowe A, Obeiro J, Newbold CI, Marsh K. Plasmodium falciparum rosetting is associated with malaria severity in Kenya. Infect Immun. 1995; 63:2323–2326.

38. Xiang L, Rundles JR, Hamilton DR, Wilson JG. Quantitative alleles of CR1: coding sequence analysis and comparison of haplotypes in two ethnic groups. J Immunol. 1999; 163:4939–4945.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download