Abstract
Purpose
The association of interleukin-10 (IL-10) polymorphisms (-1082G/A, -819C/T, -592A/C) and interleukin-6 (IL-6) poly-morphisms (-174G/C) with tuberculosis (TB) risk has been widely reported. However, the results are controversial. To clarify the role of these polymorphisms in TB, we performed a meta-analysis of all available and relevant published studies.
Materials and Methods
Based on comprehensive searches of the PubMed, Medline, Embase, Web of Science, Elsevier Science Direct and Cochrane Library database, we identified outcome data from all articles estimating the association between IL-10 and IL-6 polymorphisms and TB risk.
Results
The results indicated significant association of the allele model, heterozygous model and dominant model of IL-6 -174G/C polymorphism with decreased risk of TB. In the stratified analysis by ethnicity, significantly increased risk was observed for IL-10 -1082G/A polymorphism in Europeans under recessive model, for IL-10 -819C/T polymorphism in Asians under heterozygous model and dominant model and IL-10 -592A/C polymorphism in Asians under Allele model, homozygous model and recessive model. Moreover, significantly decreased risk of TB was associated with Asians for IL-6 -174C/G polymorphism in allele model, heterozygous model and dominant model. We also performed the analyses by sample types in IL-10 -1082G/A polymorphism, and observed significantly increased TB risk in mixed group under homozygous model.
Tuberculosis (TB) is one of the important leading causes of death in humans, and it remains a serious public health obstacle in the developing countries. It is estimated that 1.4 million people annually die due to this treatable disease and 9 million incident cases of TB are estimated globally.1 According to the report, Mycobacterium tuberculosis (MTB) infect about one-third of population; however, only approximately one-tenth of those infected will ever develop active TB, which indicate that MTB infection is the result of the interplay between host genetic susceptibility and environmental factors.2
Interleukin-10 (IL-10) is a multifunctional regulatory cytokine of inflammatory responses. Increasing numbers of studies3 have demonstrated that IL-10 acts as a general inhibitor of proliferative and cytokine responses of both T helper (Th) 1 and Th2 cells in vitro and in vivo. IL-10 plays an anti-inflammatory action by suppressing the production of cytokines such as IL-1α, IL-1β, IL-6, IL-8, IL-12, and tumor necrosis factor-alpha in activated macrophage and interferon gamma in T cells. The IL-10 gene is located on chromosome 1 (1q31-1q32) with five exons. The promoter region of IL-10 gene has been found to be highly polymorphic and its many polymorphisms have been identified.4 In the past few years, the impact of three common polymorphisms in the promoter of IL-10 gene -592A/C, -1082 G/A, and -819C/T on susceptibility to TB have been reported, and results suggested that these polymorphisms contribute to the risk of TB by affecting IL-10 transcription level, but the findings are controversial.
The human interleukin-6 (IL-6) gene is located at 7p21-24 locus with an upstream promoter containing 303 bp. IL-6 is a pleiotropic cytokine, secreted as a T-cell derived factor by a variety of cell types including lymphocytes, monocytes, and endothelial cells. It has endocrine as well as paracrine and autocrine actions implicated in several physiologic and pathologic processes including immunity and inflammation, activation of fibroblasts, mast cells, endothelial cells, monocytes, and keratinocytes.5 Furthermore, the genetic polymorphism in the IL-6 promoter (-174G/C) that influences its transcription rate might play a crucial role in host immunity and susceptibility to TB.6
A relatively large number of studies found the association between IL-10 and IL-6 polymorphisms and TB risk, however, the results have been inconsistent and inconclusive due to limited sample sizes and different study populations. Therefore, we performed this meta-analysis on all eligible case-control studies to estimate the effect of polymorphisms in the IL-10 and IL-6 genes on the risk of TB.
Relevant publications were identified with a literature search using terms "IL-10" or "Interleukin-10" or "IL-6" or "Interleukin-6" and "tuberculosis" or "TB" or "TB infection" or "TB disease" and "polymorphism" or "genotype" or "variant" in the PubMed, Medline, Embase, Web of Science, Elsevier Science Direct and Cochrane Library database (the last search update was 1 February 2014), and the search was limited to English-language journals. Additional studies were identified by a manual search of the references of original studies. The following criteria were used for inclusion in the analysis: 1) a case-control or cohort design was used and 2) studies contained available genotype frequencies. The major reasons for exclusion of studies were: no usable data were reported.
Two investigators independently extracted data and jointly reached a consensus on all of the studies researched. The following data were collected from each study: first author's name, publication year, original country, ethnicity, number of cases and controls, genotype frequencies for cases and controls, Hardy-Weinberg equilibrium (HWE) of controls and Newcastle-Ottawa Scale (NOS).7 Star symbol was used to denote the quality, based on 3 aspects of the study: selection, comparability, and exposure. Studies with a score of 7 stars or greater were considered to be of high quality.
The risks [odds ratios (ORs), and 95% confidence intervals (95% CIs)] of TB associated with IL-10 and IL-6 polymorphisms were estimated for each study based on extracted genotype data. The statistical significance of the pooled OR was determined using the Z-test. Heterogeneity assumption was examined by the Cochran's Q-test. If Q-test indicated p<0.10, thus indicating a lack of heterogeneity among studies, then the fixed effect model was used (the Mantel-Haenszel method).8 Otherwise, the random-effects model (the DerSimonian and Laird method)9 was performed. Sensitivity analysis was mainly performed to assess the stability of the results, namely, a single study in the meta-analysis was deleted to reflect the influence of the individual data set on the pooled OR. Asymmetry funnel plots were inspected to assess potential publication bias. The Egger's linear regression test was also used to assess publication bias statistically. All the above statistical analyses were performed by using the software Stata Version 12.0 (Stata Corporation, College Station, TX, USA) and p values were two-tailed.
The flow chart that displays the study selection process is shown in Fig. 1. The search of the selected databases retrieved 30 potentially relevant articles, including 7800 cases and 8793 controls, according to inclusion and exclusion criteria. There are 26 case-control studies concerning IL-10 -1082G/A polymorphism,610111213141516171819202122232425262729303132333435 15 case-control studies for IL-10 -819C/T polymorphism,61014161720212224252829303236 16 case-control studies for IL-10 -592A/C polymorphism,6101415161720212425262930323336 and 7 case-control studies about IL-6 -174G/C polymorphism. 6161721253738 Among the 30 eligible studies, 14 of them were of Asians,611151819212223283031353738 6 studies were of Europeans,121316202534 6 studies were of Africans,101424272936 and 4 studies were of Americans.17263233 The NOS scores ranged from 7 to 9, indicating that the methodological quality was generally good. The detailed characteristics of the eligible studies included in this meta-analysis are shown in Table 1, and the genotype and allele distributions of all four polymorphisms are shown in Table 2. The genotype distributions among the controls of all studies were consistent with the HWE except for eight studies for the IL-10 -1082G/A,611132325272934 one study for the IL-10 -819C/T,36 three studies for the IL-10 -592A/C,143336 and two studies for the IL-6 -174G/C (Table 1).616
The summary of the meta-analysis for IL-10 -1082G/A, -819C/T, -592A/C, and IL-6 -174G/C polymorphisms and tuberculosis susceptibility is shown in Table 3.
In all, twenty-six studies consisted of 5949 cases and 6948 controls, and assessed the potential influence of the IL-10 -1082G/A polymorphism with TB susceptibility. Random effects models were used to calculate the pooled OR in all genetic models. Overall, the combined results showed no significant association in all genetic models (Fig. 2A-E). In the stratified analysis by ethnicity, IL-10 -1082G/A polymorphism was associated with a significantly increased risk of TB in European group under recessive model (GG vs. AG+AA: OR=1.69, 95% CI=1.19-2.39). However, no significant association was found in American, Asian and African populations in all tested models. On subgroup analysis by sample types, significantly increased TB risk was observed under homozygous model (GG vs. AA: OR= 2.00, 95% CI=1.16-3.45) in PTB and extra-pulmonary tuberculosis (EPTB) mixed group. The results are shown in Table 3.
As for IL-10 -819C/T, there were fifteen studies involving 4207 cases and 5264 controls for data synthesis in our meta-analysis. The results showed that IL-10 -819C/T polymorphism was not significantly associated with the risk of TB in all genetic models (Fig. 2 F-J ). In the stratified analyses by ethnicity and control source for the -819C/T polymorphism, a significantly increased risk was observed among Asians in heterozygous model and dominant model (TC vs. CC: OR=1.34, 95% CI=1.02-1.77; TT+TC vs. CC: OR=1.31, 95% CI=1.01-1.70). The results are shown in Table 3.
In total, sixteen studies including 4115 cases and 5441 controls examined the relationship between the IL-10 -592A/C polymorphism and TB susceptibility. As shown in Table 3, we failed to find the association between the IL-10 -592A/C polymorphism and TB risk in all genetic models. In the stratified analyses for the IL-10 -592A/C polymorphism, a significantly increased risk was observed among Asians in allele model (A allele vs. C allele: OR=1.26, 95% CI=1.08-1.28), homozygous model (AA vs. CC: OR=1.50, 95% CI=1.07-2.12), and recessive model (AA vs. AC+CC: OR=1.33, 95% CI=1.10-1.62) (Table 3).
A total of 1138 cases and 1311 controls from seven case-control studies were included for data synthesis. A decreased risk between IL-6 -174G/C polymorphism and the risk of TB was observed in Allele model (C allele vs. G allele: OR=0.77, 95% CI=0.64-0.91), heterozygous model (CC vs. GG: OR=0.72, 95% CI=0.57-0.90), and dominant genetic model (CC+CG vs. GG: OR=0.71, 95% CI=0.57-0.88). In the stratified analysis by ethnicity, IL-6 -174G/C polymorphism was associated with a significantly decreased risk of TB in Asian populations in Allele model (C allele vs. G allele: OR=0.71, 95% CI=0.54-0.93), heterozygous model (CC vs. GG: OR=0.61, 95% CI=0.44-0.85), and dominant genetic model (CC+CG vs. GG: OR=0.63, 95% CI=0.46-0.86). The results are shown in Table 3.
There were statistically significant heterogeneity in all genetic models for IL-10 -1082G/A polymorphism, heterozygous model and dominant model for IL-10 -819C/T polymorphism, and all genetic models except for heterozygous model for IL-10 -592A/C (Table 3). To elucidate the heterogeneity, Galbraith plots were performed in these genetic models. When the studies which were outliers in some genetic models were excluded respectively, all I2 values were less than 50%, and Pheterogeneity were greater than 0.1 (Fig. 3, Table 4). The significance of pooled OR in all genetic models was not influenced after excluding the studies. By meta-regression analysis, the heterogeneity sources were attributable to the sample types, ethnicity, control source, and the genotyping method. Ethnicity and sample types might be predominant sources of heterogeneity in IL-10 -1082G/A polymorphism, and ethnicity and control source in both IL-10 -819C/T and IL-10 -592A/C polymorphisms (Table 5).
Sensitivity analysis was performed by sequentially excluding individual studies, including studies which was not in agreement with HWE. Statistically similar results were obtained in all genetic models after sequentially excluding each study, indicating the stability of our data.
Begg's funnel plot and Egger's test were performed to assess the publication bias of included studies. The shapes of the funnel plots did not reveal any evidence of obvious asymmetry in the all genetic models. In all genetic models, Egger's test also did not show any significant statistical evidence of publication bias, indicating low risk of publication bias in this meta-analysis (Fig. 4, Table 6).
This is not the first meta-analysis to assess the associations between three polymorphisms (-1082G/A, -819C/T, and -592A/C) in the IL-10 gene promoter and the risk of TB. We found that the results of our meta-analysis are inconsistent with a recent study of Liang, et al.39 in which some following shortcomings were found : 1) the NOS scores of 3 Chinese articles included were lower than 7 stars through quality assessment, 2) two studies that meet the inclusion criterion were excluded (Ma, et al.,28 Spinassé, et al.32), 3) the choice of genetic models was incorrect, 4) heterogeneity analysis and sensitivity analysis were missing, and 5) some extracted data was not accurate enough. Therefore, we performed this meta-analysis to examine the association between three IL-10 and IL-6 polymorphisms and TB risk again. Our meta-analysis results indicated that the presence of the IL-10 -1082G/A, -819C/T, and -592A/C polymorphisms was not associated with the risk of TB in all genetic models. On the other hand, the IL-6 -174G/C polymorphism might be associated with an decreased risk of TB in some genetic models (C allele vs. G allele: OR=0.77, 95% CI=0.64-0.91, p=0.003; CC vs. GG: OR=0.72, 95% CI=0.57-0.90, p=0.005; CC+CG vs. GG: OR=0.71, 95% CI=0.57-0.88, p=0.002).
We also carried out subgroup analysis based on ethnicity, sample types and control source in consideration of obvious heterogeneity. In the stratiied analysis by ethnicity, we observed significantly increased TB risk associated with the IL-10 -1082G/A polymorphism in recessive model in Europeans, IL-10 -819C/T polymorphism in Asians in heterozygous model and dominant model, IL-10 -592A/C polymorphism in Asians in Allele model, homozygous model and recessive model respectively, and a decreased TB risk associated with IL-6 -174G/C polymorphism was found in allele model, heterozygous model and dominant model in Asians. Different genetic background and environmental exposures might contribute to this ethnic difference. Subgroup analysis based on sample types suggested that IL-10 -1082G/A polymorphism may be related with an increased risk of TB in homozygous model in the PTB+EPTB mixed sample. The results of subgroup analysis by control source revealed no signiicant association with TB susceptibility among IL-10 and IL-6 polymorphisms.
In our meta-analysis, obvious heterogeneity was observed for IL-10 -1082G/A polymorphism in all genetic models, -819C/T polymorphism in heterozygous model and dominant model, and -592A/C polymorphism in all genetic models except for heterozygous model, whereas there was no obvious heterogeneity for IL-6 -174G/C polymorphism. Then, we used the Galbraith plots to explore the sources of heterogeneity. We found that all the I2 values were less than 50% and Pheterogeneity were greater than 0.1 after excluding some studies, thus indicating that these studies might be the major source of the heterogeneity for the IL-10 -1082G/A, -819C/T, and -592A/C polymorphisms. Owing to the limited number of studies in this meta-analysis, we restricted meta-regression analysis to four factors (sample types, ethnicity, control source, and genotyping method), which are the most likely to cause the heterogeneity between studies. Although the four above-mentioned factors had no significant impact on the heterogeneity except sample types factor for IL-10 -1082G/A in homozygous model, the results of subgroup analyses revealed that the ethnicity and sample type might contribute to the potential heterogeneity.
Some limitations of this meta-analysis exist which should be considered when interpreting the present results. Firstly, heterogeneity is a potential problem when interpreting the results of meta-analysis. Significant heterogeneity existed among some comparisons, especially for IL-10 -1082G/A and -592A/C polymorphisms. Secondly, this meta-analysis included the only published studies and publication bias may occur, although our results of publication bias showed no significance. Thirdly, host genetic susceptibility, environment factors and other factors might contribute to the pathogenesis of TB. Although many other factors such as age or gender may play a profound role in the development of TB, we did not make subgroup analysis based on these factors as data is not sufficient. Finally, some genetic polymorphisms of studies deviant from HWE were included in this meta-analysis, which suggested that there was potential bias during control selection or genotyping errors.
In conclusion, our meta-analysis suggested that IL-10 -1082G/A, -819C/T, and -592A/C polymorphisms had no association with TB risk in general population, while the IL-6 -174G/C polymorphism was signiicantly associated with decreased risk of TB in all genetic models except for recessive model. In the subgroup analysis, IL-10 -1082G/A polymorphism was associated with TB risk in Europeans in recessive model, and IL-10 -592A/C polymorphisms were significantly associated with TB risk in Asians in Allele model, homozygous model and recessive model, respectively, and a decreased TB risk associated with IL-6 -174G/C polymorphism was found in allele model, heterozygous model and dominant model in Asians. Furthermore, IL-10 -1082G/A polymorphism was associated also with an increased risk of TB in homozygous model in the PTB+EPTB mixed sample. However, additional well-designed and larger scale primary studies in populations with different ethnicities are required to further evaluate the IL-10 and IL-6 gene polymorphisms with TB risk in future.
Figures and Tables
 | Fig. 2Forest plot of the overall risk of TB associated with the IL-10 -1082G/A and -819C/T polymorphism in all genetic models. Bars represent 95% CI and boxes represent OR values. The size of each box indicates the weight of the study in the pooled results. (F-J) T allele vs. C allele, TT vs. CC, TC vs. CC, TT+TC vs. CC, TT vs. TC+CC for -819C/T. TB, tuberculosis; IL-10, interleukin 10; CI, confidence interval; OR, odds ratio. |
 | Fig. 3Galbraith plot of IL-10 promoter polymorphism and TB risk. (A-E) The five studies1820223133 in G vs. A, three studies202231 in GG vs. AA, seven studies13182022313337 in AG vs. AA, six studies131820223133 in GG+AG vs. AA, and five studies2022273537 in GG vs. AG+AA were outliers for -1082G/A. (F and G) The one study38 in TC vs. CC and one study38 in TT+TC vs. CC for -819C/T. (H-K) The three studies262838 in A vs. C, one study38 in AA vs. CC, one study38 in AA+AC vs. CC, and two studies1738 in AA vs. AC+CC for -592A/C. TB, tuberculosis; IL-10, interleukin 10. |
 | Fig. 4Funnel plot for publication bias of the meta-analysis of tuberculosis risk and IL-10 polymorphisms in allele genetic model comparison. (A) IL-10 -1082G/A polymorphism. (B) IL-10 -819C/T polymorphism. (C) IL-10 -592A/C polymorphism. (D) IL-6 -174G/C polymorphism. IL-10, interleukin 10; IL-6, interleukin 6. |
Table 1
Baseline Characteristics of the 30 Eligible Studies Included in This Meta-Analysis

| Study | Yr | Male patients (%) | Mean age (yrs) | Sample types | Sample size | SNP studied | Clinical diagnoses performed | Control source | Sample tested | Genotyping method | NOS score | P-HWE for controls | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Control | |||||||||||
| IL-10 | ||||||||||||||
| Bellamy, et al.10 | 1998 | 67.4 | 34.7±13.2 | 30.3±7.5 | PTB | 401 | 408 | -1082G/A, -819C/T, -592A/C | Acidfast, bacilli (AFB) | HB | Blood | PCR-slot-blotting | 7 | 0.824 |
| Delgado, et al.11 | 2002 | 37.3 | 42.2±14.1 | 37.5±12.9 | PTB | 356 | 106 | -1082G/A | Sputum smear, medical history, physical examination | HB | Blood | RFLP-PCR | 9 | <0.001 |
| López-Maderuelo, et al.12 | 2003 | NR | NR | NR | PTB | 113 | 100 | -1082G/A | Culture, radiologic diagnosed | HB | Blood | ARMS-PCR | 8 | 0.949 |
| Scola, et al.13 | 2003 | NR | 35-60 | NR | PTB | 45 | 114 | -1082G/A | Clinical history, radiologic diagnosed | PB | Blood | ARMS-PCR | 7 | <0.001 |
| Fitness, et al.14 | 2004 | NR | NR | NR | PTB | 210 | 705 | -1082G/A, -819C/T, -592A/C | Culture, smear, history | HB | Blood | ARMS-PCR | 7 | 0.524 |
| Shin, et al.15 | 2005 | NR | 46.9 (18-86) | 56.1 (50-81) | PTB | 450 | 851 | -1082G/A, -592A/C | AFB | HB | Blood | Single-base extension methods | 8 | 0.168 |
| Amirzargar, et al.6 | 2006 | NR | NR | NR | PTB | 41 | 123 | -1082G/A, -819C/T, -592A/C | AFB, chest X-ray (CXR) | HB | Blood | PCR-SSP | 8 | <0.001 |
| Oral, et al.16 | 2006 | NR | NR | NR | PTB, EPTB | 81 | 50 | -1082G/A, -819C/T, -592A/C | Staining of sputum smears, culture, biopsy, radiography | HB | Blood | PCR-SSP | 9 | 0.06 |
| Henao, et al.17 | 2006 | 57.9 | 15-70 | 17-55 | PTB, EPTB | 190 | 135 | -1082G/A, -819C/T, -592A/C | Ziehl-Nielssen staining of sputum smears, culture, biopsy, CXR, clinical history | HB | Blood | PCR-SSP | 9 | 0.94 |
| Oh, et al.18 | 2007 | 68.9 | 17-88 | 45.8 (18-81) | PTB | 145 | 117 | -1082G/A | Staining of sputum smears, culture, radiography | HB | Blood | ARMS-PCR | 8 | 0.612 |
| Prabhu Anand, et al.19 | 2007 | 56.8 | 35.5±12.3 | 29.7±9.5 | PTB | 132 | 143 | -1082G/A | Staining of sputum smears, culture, radiography | HB | Blood | PCR-RFLP | 8 | 0.123 |
| Ates, et al.20 | 2008 | 62 | 47.84±12.6 | 54.1±7.2 | PTB, EPTB | 128 | 80 | -1082G/A, -819C/T, -592A/C | Radiographic, clinical presentation, smears, culture | HB | Blood | ARMS-PCR | 9 | 0.978 |
| Selvaraj, et al.21 | 2008 | 71.7 | Male: 35.3±10.5, female: 29.2±10.3 | Male: 32±8.1, female: 27.1±8.6 | PTB | 155 | 183 | -1082G/A, -819C/T, | Radiographic, clinical presentation, smears, culture | PB | Blood | PCR-RFLP | 7 | 0.204 |
| Wu, et al.22 | 2008 | NR | NR | NR | PTB | 61 | 122 | -1082G/A, -819C/T, -592A/C | Radiographic, clinical presentation, smears, culture | HB | Blood | PCR-RFLP | 7 | 0.379 |
| Ansari, et al.23 | 2009 | NR | NR | NR | PTB | 188 | 188 | -1082G/A | Microscopy, culture, histology, imaging | PB | Blood | ARMS-PCR | 8 | <0.001 |
| Thye, et al.24 | 2009 | NR | NR | NR | PTB | 2010 | 2346 | -1082G/A, -819C/T, -592A/C | Smears, culture | PB | Blood | FRET | 8 | 0.542 |
| Trajkov, et al.25 | 2009 | NR | 20-59 | NR | PTB | 75 | 299 | -1082G/A, -819C/T, -592A/C | WHO based | PB | Blood | PCR-SSP | 7 | <0.001 |
| Taype, et al.26 | 2010 | 97.6 | 29.01±11.42 | 32.56±9.39 | PTB, EPTB | 626 | 513 | -1082G/A, -592A/C | Smears, culture, biopsy, clinical | HB | Blood | Taqman PCR | 9 | 0.142 |
| IL-10 | ||||||||||||||
| Mosaad, et al.27 | 2010 | 67.3 | 0.5 (0.025-1.5) | NR (0.047-1.5) | PTB, EPTB | 110 | 118 | -1082G/A | Smear, culture | HB | Blood | ARMS-PCR | 9 | <0.001 |
| Ma, et al.28 | 2010 | 27.8 | 34.75±16.67 | 38.17±17.39 | PTB | 543 | 544 | -819C/T | Radiographic, smears, culture | HB | Blood | ARMS-PCR | 9 | 0.491 |
| Ben-Selma, et al.29 | 2011 | 51.9 | PTB: 44, EPTB: 39 | 35 | PTB, EPTB | 131 | 95 | -1082G/A, -819C/T, -592A/C | Sputum smear, CXR, radiologic, histologic grounds | HB | Blood | PCR-RFLP | 9 | <0.05 |
| Liang, et al.30 | 2011 | NR | NR | NR | PTB, EPTB | 235 | 78 | -1082G/A, -819C/T, -592A/C | Radiographic, biopsy, clinical presentation, smears, culture | HB | Blood | SNaPshot assay | 9 | 0.589 |
| Ramaseri Sunder, et al.31 | 2012 | NR | NR | NR | PTB, EPTB | 104 | 102 | -1082G/A | Sputum smear, CXR, biopsy, Fine Needle Aspiration Cytology (FNAC) | HB | Blood | ARMS-PCR | 8 | 0.057 |
| Spinassé, et al.32 | 2012 | NR | NR | NR | PTB | 221 | 271 | -1082G/A, -819C/T, -592A/C | Culture | HB | Blood | Sequencing | 7 | 0.189 |
| García-Elorriaga, et al.33 | 2013 | 38.9 | 38-65 | 26-41 | PTB | 77 | 60 | -1082G/A, -592A/C | WHO based | HB | Blood | Taqman PCR | 7 | 0.728 |
| Ulger, et al.34 | 2013 | 84.5 | 32.57±15.94 | 29.40±11.56 | PTB, EPTB | 84 | 110 | -1082G/A | Smear, culture | HB | Blood | PCR-RFLP | 8 | <0.001 |
| Meenakshi, et al.35 | 2013 | 50 | 27.4±13.9 | 30±10.7 | PTB | 100 | 100 | -1082G/A | Radiographic, sputum culture, AFB, histocytological examination | HB | Blood | ARMS-PCR | 8 | 0.058 |
| Mhmoud, et al.36 | 2013 | 69.6 | 36.9 (15-89) | 31.2 (17-85) | PTB | 191 | 206 | -819C/T, -592A/C | Culture, smear | HB | Blood | PCR-RFLP | 8 | <0.001 |
| IL-6 | ||||||||||||||
| Oral, et al.16 | 2006 | NR | NR | NR | PTB, EPTB | 81 | 49 | -174G/C | Staining of sputum smears, culture, biopsy, radiography | HB | Blood | PCR-SSP | 9 | <0.05 |
| Henao, et al.17 | 2006 | 57.9 | 15-70 | 17-55 | PTB, EPTB | 190 | 135 | -174G/C | Ziehl-Nielssen staining of sputum smears, culture, biopsy, CXR, clinical history | HB | Blood | PCR-SSP | 9 | 0.689 |
| Amirzargar, et al.6 | 2006 | NR | NR | NR | PTB | 40 | 119 | -174G/C | AFB, CXR | HB | Blood | PCR-SSP | 8 | <0.05 |
| Selvaraj, et al.21 | 2008 | 71.7 | Male: 35.3±10.5, female: 29.2±10.3 | Male: 32±8.1, female: 27.1±8.6 | PTB | 160 | 183 | -174G/C | Radiographic, clinical presentation, smears, culture | PB | Blood | PCR-RFLP | 7 | 0.419 |
| Trajkov, et al.25 | 2009 | NR | 20-59 | NR | PTB | 75 | 301 | -174G/C | WHO based | PB | Blood | PCR-SSP | 7 | 0.492 |
| Ansari, et al.37 | 2011 | NR | Minimal/moderate disease: 32.4±15.9; advanced disease: 27±17.0 | 28.3±12.1 | PTB | 97 | 166 | -174G/C | Radiographic, smears, culture | PB | Blood | ARMS-PCR | 8 | 0.567 |
| Zhang, et al.38 | 2012 | 62 | 38.64±18.44 | 36.92±16.52 | PTB | 495 | 358 | -174G/C | Radiographic, smears, culture | HB | Blood | Mass spectrometry | 8 | 0.979 |
NR, not report; PTB, pulmonary tuberculosis; EPTB, extra-pulmonary tuberculosis; SNP, single nucleotide polymorphism; PB, population-based controls; HB, hospital-based controls; PCR, polymerase chain reaction; SSP, sequence-specific primers; ARMS, amplification refractory mutation system; RFLP, restriction fragment length polymorphism; NOS, newcastle-ottawa scale; C, confirmed to HWE; HWE, Hardy-Weinberg equilibrium.
Table 2
Genotype and Allele Distributions of IL-10 and IL-6 Polymorphisms in Cases and Controls

| Polymorphisms | Study | Country | Ethnicity | Case | Control | Case | Control | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | AG | AA | GG | AG | AA | G | A | G | A | ||||
| IL-10-1082G/A | Bellamy, et al.10 | Gambia | African | 51 | 185 | 165 | 45 | 184 | 179 | 287 | 515 | 274 | 542 |
| Delgado, et al.11 | Cambodia | Asian | 11 | 259 | 86 | 3 | 64 | 39 | 281 | 431 | 70 | 142 | |
| López-Maderuelo, et al.12 | Spain | European | 33 | 47 | 33 | 29 | 50 | 21 | 113 | 113 | 108 | 92 | |
| Scola, et al.13 | Italy | European | 17 | 22 | 6 | 24 | 77 | 13 | 56 | 34 | 125 | 103 | |
| Fitness, et al.14 | Malawi | African | 23 | 78 | 69 | 87 | 251 | 203 | 124 | 216 | 425 | 657 | |
| Shin, et al.15 | Korea | Asian | 2 | 53 | 394 | 9 | 124 | 718 | 57 | 841 | 142 | 1560 | |
| Amirzargar, et al.6 | Iran | Asian | 2 | 31 | 7 | 5 | 79 | 18 | 35 | 45 | 89 | 115 | |
| Oral, et al.16 | Turkey | European | 10 | 41 | 30 | 5 | 13 | 32 | 61 | 101 | 23 | 77 | |
| Henao, et al.17 | Colombia | American | 32 | 92 | 66 | 26 | 66 | 43 | 156 | 224 | 118 | 152 | |
| Oh, et al.18 | Korea | Asian | 4 | 43 | 98 | 19 | 53 | 45 | 51 | 239 | 91 | 143 | |
| Prabhu, et al.19 | India | Asian | 3 | 55 | 74 | 6 | 61 | 73 | 61 | 203 | 73 | 207 | |
| Ates, et al.20 | Turkey | European | 26 | 65 | 37 | 6 | 32 | 42 | 117 | 139 | 44 | 116 | |
| Selvaraj, et al.21 | India | Asian | 5 | 42 | 102 | 6 | 69 | 108 | 52 | 246 | 81 | 285 | |
| Wu, et al.22 | China | Asian | 1 | 12 | 48 | 0 | 18 | 104 | 14 | 108 | 18 | 226 | |
| Ansari, et al.23 | Pakistan | Asian | 27 | 132 | 29 | 20 | 136 | 32 | 186 | 190 | 176 | 200 | |
| Thye, et al.24 | Ghana | African | 117 | 631 | 793 | 160 | 783 | 1025 | 865 | 2217 | 1103 | 2833 | |
| Trajkov, et al.25 | Macedonia | European | 10 | 48 | 17 | 17 | 212 | 70 | 68 | 82 | 246 | 352 | |
| Taype, et al.26 | Peru | American | 22 | 187 | 414 | 10 | 153 | 347 | 231 | 1015 | 173 | 847 | |
| Mosaad, et al.27 | Egypt | African | 16 | 92 | 2 | 22 | 88 | 8 | 124 | 96 | 132 | 104 | |
| Ben-Selma, et al.29 | Tunisian | African | 21 | 65 | 45 | 9 | 26 | 60 | 168.8 | 155 | 44 | 146 | |
| Liang, et al.30 | China | Asian | 0 | 28 | 207 | 0 | 9 | 69 | 28 | 442 | 9 | 147 | |
| Ramaseri Sunder, et al.31 | India | Asian | 3 | 25 | 76 | 2 | 43 | 57 | 31 | 177 | 47 | 157 | |
| Spinassé, et al.32 | Brazil | American | 24 | 100 | 97 | 31 | 107 | 133 | 148 | 294 | 169 | 373 | |
| García-Elorriaga, et al.33 | Mexico | American | 54 | 20 | 3 | 31 | 25 | 4 | 128 | 26 | 87 | 33 | |
| Ulger, et al.34 | Turkey | European | 0 | 84 | 0 | 1 | 104 | 5 | 84 | 84 | 106 | 114 | |
| Meenakshi, et al.35 | India | Asian | 4 | 81 | 15 | 16 | 59 | 25 | 89 | 111 | 91 | 109 | |
| Polymorphisms | Study | Country | Ethnicity | Case | Control | Case | Control | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | TC | CC | TT | TC | CC | T | C | T | C | ||||
| IL-10-819C/T | Bellamy, et al.10 | Gambia | African | 89 | 192 | 120 | 88 | 206 | 114 | 370 | 432 | 382 | 434 |
| Fitness, et al.14 | Malawi | African | 27 | 98 | 85 | 108 | 303 | 287 | 152 | 268 | 519 | 877 | |
| Amirzargar, et al.6 | Iran | Asian | 2 | 20 | 19 | 9 | 52 | 62 | 24 | 58 | 70 | 176 | |
| Oral, et al.16 | Turkey | European | 10 | 23 | 48 | 7 | 19 | 24 | 43 | 119 | 33 | 67 | |
| Henao, et al.17 | Colombia | American | 32 | 92 | 66 | 21 | 64 | 50 | 156 | 224 | 106 | 164 | |
| Ates, et al.20 | Turkey | European | 7 | 58 | 63 | 8 | 36 | 36 | 72 | 184 | 52 | 108 | |
| Selvaraj, et al.21 | India | Asian | 45 | 86 | 24 | 56 | 82 | 45 | 176 | 134 | 194 | 172 | |
| Wu, et al.22 | China | Asian | 24 | 34 | 3 | 50 | 62 | 10 | 82 | 40 | 162 | 82 | |
| Thye, et al.24 | Ghana | African | 267 | 762 | 515 | 365 | 942 | 665 | 1296 | 1792 | 1672 | 2272 | |
| Trajkov, et al.25 | Macedonia | European | 5 | 35 | 35 | 19 | 125 | 155 | 45 | 105 | 163 | 435 | |
| Ma, et al.28 | China | Asian | 229 | 256 | 58 | 230 | 253 | 61 | 714 | 372 | 713 | 375 | |
| Ben-Selma, et al.29 | Tunisian | African | 11 | 65 | 55 | 10 | 42 | 43 | 87 | 175 | 62 | 128 | |
| Liang, et al.30 | China | Asian | 123 | 90 | 22 | 35 | 31 | 12 | 336 | 134 | 101 | 55 | |
| Spinassé, et al.32 | Brazil | American | 32 | 100 | 89 | 38 | 124 | 109 | 164 | 278 | 200 | 342 | |
| Mhmoud, et al.36 | Sudan | African | 42 | 126 | 23 | 70 | 73 | 63 | 210 | 172 | 213 | 199 | |
| Polymorphisms | Study | Country | Ethnicity | Case | Control | Case | Control | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AC | CC | AA | AC | CC | A | C | A | C | ||||
| IL-10-592A/C | Bellamy, et al.10 | Gambia | African | 89 | 192 | 120 | 88 | 206 | 114 | 370 | 432 | 382 | 434 |
| Fitness, et al.14 | Malawi | African | 27 | 98 | 85 | 107 | 301 | 297 | 152 | 268 | 515 | 895 | |
| Shin, et al.15 | Korea | Asian | 238 | 173 | 39 | 376 | 384 | 91 | 649 | 251 | 1136 | 566 | |
| Amirzargar, et al.6 | Iran | Asian | 2 | 20 | 18 | 9 | 52 | 62 | 24 | 56 | 70 | 176 | |
| Polymorphisms | Study | Country | Ethnicity | Case | Control | Case | Control | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AC | CC | AA | AC | CC | A | C | A | C | ||||
| Oral, et al.16 | Turkey | European | 10 | 23 | 48 | 7 | 19 | 24 | 43 | 119 | 33 | 67 | |
| Henao, et al.17 | Colombia | American | 72 | 89 | 29 | 41 | 67 | 27 | 233 | 147 | 149 | 121 | |
| Ates, et al.20 | Turkey | European | 7 | 58 | 63 | 8 | 36 | 36 | 72 | 184 | 52 | 108 | |
| Wu, et al.22 | China | Asian | 24 | 34 | 3 | 50 | 62 | 10 | 82 | 40 | 162 | 82 | |
| Thye, et al.24 | Ghana | African | 172 | 532 | 321 | 269 | 696 | 480 | 876 | 1174 | 1234 | 1656 | |
| Trajkov, et al.25 | Macedonia | European | 5 | 31 | 39 | 28 | 117 | 154 | 41 | 109 | 173 | 425 | |
| Taype, et al.26 | Peru | American | 117 | 218 | 264 | 105 | 230 | 178 | 452 | 746 | 440 | 586 | |
| Ben-selma, et al.29 | Tunisian | African | 12 | 63 | 56 | 10 | 42 | 43 | 87 | 175 | 62 | 128 | |
| Liang, et al.30 | China | Asian | 123 | 90 | 22 | 35 | 31 | 12 | 336 | 134 | 101 | 55 | |
| Spinassé, et al.32 | Brazil | American | 34 | 102 | 85 | 37 | 127 | 107 | 170 | 272 | 201 | 341 | |
| García-elorriaga, et al.33 | Mexico | American | 1 | 57 | 19 | 1 | 42 | 17 | 59 | 95 | 44 | 76 | |
| Mhmoud, et al.36 | Sudan | African | 127 | 47 | 17 | 100 | 68 | 38 | 301 | 81 | 268 | 144 | |
| Polymorphisms | Study | Country | Ethnicity | Case | Control | Case | Control | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CG | GG | CC | CG | GG | C | G | C | G | ||||
| IL-6-174G/C | Oral, et al.16 | Turkey | European | 6 | 27 | 48 | 9 | 13 | 27 | 39 | 123 | 31 | 67 |
| Henao, et al.17 | Colombia | American | 11 | 73 | 106 | 13 | 61 | 61 | 95 | 285 | 87 | 183 | |
| Amirzargar, et al.6 | Iran | Asian | 4 | 13 | 23 | 10 | 71 | 38 | 21 | 59 | 91 | 147 | |
| Selvaraj, et al.21 | India | Asian | 3 | 35 | 122 | 3 | 51 | 129 | 41 | 279 | 57 | 309 | |
| Trajkov, et al.25 | Macedonia | European | 8 | 31 | 36 | 25 | 132 | 144 | 47 | 103 | 182 | 420 | |
| Ansari, et al.37 | Pakistan | Asian | 4 | 24 | 69 | 10 | 56 | 100 | 32 | 162 | 76 | 256 | |
| Zhang, et al.38 | China | Asian | 0 | 4 | 491 | 0 | 1 | 357 | 4 | 986 | 1 | 715 | |
Table 3
Determination of the Genetic Effects of IL-10 and IL-6 Polymorphisms on TB and Subgroup Analysis

Table 4
Meta-Analyses of IL-10 Polymorphisms and Risk of TB after Omitting the Studies
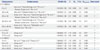
| Polymorphisms | Omitted studies | OR (95% CI) | Z | POR | I2 (%) | Pheterogeneity | Effect model |
|---|---|---|---|---|---|---|---|
| IL-10-1082G/A | |||||||
| G vs. A | Ates, et al.;20 García-Elorriaga, et al.;33 Oh, et al.;18 Ramaseri Sunder, et al.;31 Wu, et al.22 | 1.02 (0.96, 1.09) | 0.69 | 0.488 | 9.4 | 0.336 | F |
| GG vs. AA | Ates, et al.;20 Ramaseri Sunder, et al.;31 Wu, et al.22 | 1.04 (0.90, 1.21) | 0.54 | 0.588 | 2.2 | 0.430 | F |
| AG vs. AA | Ansari, et al.;37 Ates, et al.;20 García-Elorriaga, et al.;33 Oh, et al.;18 Ramaseri Sunder, et al.;31 Scola, et al.;13 Wu, et al.22 | 1.00 (0.92, 1.09) | 0.01 | 0.989 | 0.0 | 0.486 | F |
| GG+AG vs. AA | Ates, et al.;20 García-Elorriaga, et al.;33 Oh, et al.;18 Ramaseri Sunder, et al.;31 Scola, et al.;13 Wu, et al.22 | 1.01 (0.93, 1.10) | 0.25 | 0.802 | 11.6 | 0.311 | F |
| GG vs. AG+AA | Ansari, et al.;37 Ates, et al.;20 Meenakshi, et al.;35 Mosaad, et al.;27 Wu, et al.22 | 1.03 (0.90, 1.19) | 0.46 | 0.645 | 0.0 | 0.623 | F |
| IL-10-819C/T | |||||||
| TC vs. CC | Zhang, et al.38 | 1.06 (0.96, 1.17) | 1.19 | 0.234 | 0.0 | 0.671 | F |
| TT+TC vs. CC | Zhang, et al.38 | 1.04 (0.95, 1.14) | 0.80 | 0.424 | 0.0 | 0.683 | F |
| IL-10-592A/C | |||||||
| A vs. C | Ma, et al.;28 Taype, et al.;26 Zhang, et al.38 | 1.08 (0.99, 1.17) | 1.78 | 0.075 | 3.7 | 0.409 | F |
| AA vs. CC | Zhang, et al.38 | 0.99 (0.87, 1.13) | 0.11 | 0.913 | 5.9 | 0.386 | F |
| AA+AC vs. CC | Zhang, et al.38 | 1.00 (0.91, 1.10) | 0.08 | 0.936 | 29.1 | 0.138 | F |
| AA vs. AC+CC | Henao, et al.;17 Zhang, et al.38 | 0.96 (0.85, 1.09) | 0.63 | 0.526 | 0.0 | 0.845 | F |
Table 5
Multivariate Meta-Regression Analyses of Potential Source of Heterogeneity
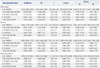
Table 6
Publication Bias of IL-10 -1082G/A, IL-10 -819C/T, and IL-10 -592A/C Polymorphisms in all Genetic Models

ACKNOWLEDGEMENTS
This work was supported by a grant from the National Clinical Key Specialty Construction Projects to the Department of Clinical Laboratory of Renmin Hospital of Wuhan University.
References
1. Gouzy A, Nigou J, Gilleron M, Neyrolles O, Tailleux L, Gordon SV. Tuberculosis 2012: biology, pathogenesis and intervention strategies; an update from the city of light. Res Microbiol. 2013; 164:270–280.

2. Hill AV. Aspects of genetic susceptibility to human infectious diseases. Annu Rev Genet. 2006; 40:469–486.

3. Abbas AK, Murphy KM, Sher A. Functional diversity of helper Tlymphocytes. Nature. 1996; 383:787–793.
4. Howell WM, Rose-Zerilli MJ. Cytokine gene polymorphisms, cancer susceptibility, and prognosis. J Nutr. 2007; 137:1 Suppl. 194S–199S.
6. Amirzargar AA, Rezaei N, Jabbari H, Danesh AA, Khosravi F, Hajabdolbaghi M, et al. Cytokine single nucleotide polymorphisms in Iranian patients with pulmonary tuberculosis. Eur Cytokine Netw. 2006; 17:84–89.
7. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in metaanalyses. Eur J Epidemiol. 2010; 25:603–605.

8. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959; 22:719–748.
10. Bellamy R, Ruwende C, Corrah T, McAdam KP, Whittle HC, Hill AV. Assessment of the interleukin 1 gene cluster and other candidate gene polymorphisms in host susceptibility to tuberculosis. Tuber Lung Dis. 1998; 79:83–89.

11. Delgado JC, Baena A, Thim S, Goldfeld AE. Ethnic-specific genetic associations with pulmonary tuberculosis. J Infect Dis. 2002; 186:1463–1468.

12. Lpez-Maderuelo D, Arnalich F, Serantes R, Gonzlez A, Codoceo R, Madero R, et al. Interferon-gamma and interleukin-10 gene polymorphisms in pulmonary tuberculosis. Am J Respir Crit Care Med. 2003; 167:970–975.

13. Scola L, Crivello A, Marino V, Gioia V, Serauto A, Candore G, et al. IL-10 and TNF-alpha polymorphisms in a sample of Sicilian patients affected by tuberculosis: implication for ageing and life span expectancy. Mech Ageing Dev. 2003; 124:569–572.

14. Fitness J, Floyd S, Warndorff DK, Sichali L, Malema S, Crampin AC, et al. Large-scale candidate gene study of tuberculosis susceptibility in the Karonga district of northern Malawi. Am J Trop Med Hyg. 2004; 71:341–349.

15. Shin HD, Park BL, Kim YH, Cheong HS, Lee IH, Park SK. Common interleukin 10 polymorphism associated with decreased risk of tuberculosis. Exp Mol Med. 2005; 37:128–132.

16. Oral HB, Budak F, Uzaslan EK, Baştürk B, Bekar A, Akalin H, et al. Interleukin-10 (IL-10) gene polymorphism as a potential host susceptibility factor in tuberculosis. Cytokine. 2006; 35:143–147.

17. Henao MI, Montes C, Pars SC, Garca LF. Cytokine gene polymorphisms in Colombian patients with different clinical presentations of tuberculosis. Tuberculosis (Edinb). 2006; 86:11–19.

18. Oh JH, Yang CS, Noh YK, Kweon YM, Jung SS, Son JW, et al. Polymorphisms of interleukin-10 and tumour necrosis factor-alpha genes are associated with newly diagnosed and recurrent pulmonary tuberculosis. Respirology. 2007; 12:594–598.

19. Prabhu Anand S, Selvaraj P, Jawahar MS, Adhilakshmi AR, Narayanan PR. Interleukin-12B & interleukin-10 gene polymorphisms in pulmonary tuberculosis. Indian J Med Res. 2007; 126:135–138.
20. Ates O, Musellim B, Ongen G, Topal-Sarikaya A. Interleukin-10 and tumor necrosis factor-alpha gene polymorphisms in tuberculosis. J Clin Immunol. 2008; 28:232–236.

21. Selvaraj P, Alagarasu K, Harishankar M, Vidyarani M, Nisha Rajeswari D, Narayanan PR. Cytokine gene polymorphisms and cytokine levels in pulmonary tuberculosis. Cytokine. 2008; 43:26–33.

22. Wu F, Qu Y, Tang Y, Cao D, Sun P, Xia Z. Lack of association between cytokine gene polymorphisms and silicosis and pulmonary tuberculosis in Chinese iron miners. J Occup Health. 2008; 50:445–454.

23. Ansari A, Talat N, Jamil B, Hasan Z, Razzaki T, Dawood G, et al. Cytokine gene polymorphisms across tuberculosis clinical spectrum in Pakistani patients. PLoS One. 2009; 4:e4778.

24. Thye T, Browne EN, Chinbuah MA, Gyapong J, Osei I, Owusu-Dabo E, et al. IL10 haplotype associated with tuberculin skin test response but not with pulmonary TB. PLoS One. 2009; 4:e5420.

25. Trajkov D, Trajchevska M, Arsov T, Petlichkovski A, Strezova A, Efinska-Mladenovska O, et al. Association of 22 cytokine gene polymorphisms with tuberculosis in Macedonians. Indian J Tuberc. 2009; 56:117–131.
26. Taype CA, Shamsuzzaman S, Accinelli RA, Espinoza JR, Shaw MA. Genetic susceptibility to different clinical forms of tuberculosis in the Peruvian population. Infect Genet Evol. 2010; 10:495–504.

27. Mosaad YM, Soliman OE, Tawhid ZE, Sherif DM. Interferon-gamma +874 T/A and interleukin-10 -1082 A/G single nucleotide polymorphism in Egyptian children with tuberculosis. Scand J Immunol. 2010; 72:358–364.

28. Ma MJ, Xie LP, Wu SC, Tang F, Li H, Zhang ZS, et al. Toll-like receptors, tumor necrosis factor-α, and interleukin-10 gene polymorphisms in risk of pulmonary tuberculosis and disease severity. Hum Immunol. 2010; 71:1005–1010.
29. Ben-Selma W, Harizi H, Boukadida J. Association of TNF-α and IL-10 polymorphisms with tuberculosis in Tunisian populations. Microbes Infect. 2011; 13:837–843.

30. Liang L, Zhao YL, Yue J, Liu JF, Han M, Wang H, et al. Interleukin-10 gene promoter polymorphisms and their protein production in pleural fluid in patients with tuberculosis. FEMS Immunol Med Microbiol. 2011; 62:84–90.

31. Ramaseri Sunder S, Hanumanth SR, Nagaraju RT, Venkata SK, Suryadevara NC, Pydi SS, et al. IL-10 high producing genotype predisposes HIV infected individuals to TB infection. Hum Immunol. 2012; 73:605–611.

32. Spinassé LB, Miranda AB, Santos AR, Mello FCQ, Lapa e Silva JR, Lopes MQP, et al. Partial Mapping of the IL-10 Promoter Region: Identification of New SNPs and Association with Tuberculosis Outcome in Brazilians. In : Cardona PJ, editor. Understanding Tuberculosis-Analyzing the Origin of Mycobacterium Tuberculosis Pathogenicity. INTECH Open Access Publisher;2012. p. 357–366.
33. Garca-Elorriaga G, Vera-Ramrez L, del Rey-Pineda G, Gonzlez-Bonilla C. -592 and -1082 interleukin-10 polymorphisms in pulmonary tuberculosis with type 2 diabetes. Asian Pac J Trop Med. 2013; 6:505–509.

34. Ulger M, Emekdas¸ G, Aslan G, Tas¸ D, Ilvan A, Tezcan S, et al. [Determination of the cytokine gene polymorphism and genetic susceptibility in tuberculosis patients]. Mikrobiyol Bul. 2013; 47:250–264.

35. Meenakshi P, Ramya S, Shruthi T, Lavanya J, Mohammed HH, Mohammed SA, et al. Association of IL-1β +3954 C/T and IL-10-1082 G/A cytokine gene polymorphisms with susceptibility to tuberculosis. Scand J Immunol. 2013; 78:92–97.

36. Mhmoud N, Fahal A, van de Sande WJ. Association of IL-10 and CCL5 single nucleotide polymorphisms with tuberculosis in the Sudanese population. Trop Med Int Health. 2013; 18:1119–1127.

37. Ansari A, Hasan Z, Dawood G, Hussain R. Differential combination of cytokine and interferon-γ +874 T/A polymorphisms determines disease severity in pulmonary tuberculosis. PLoS One. 2011; 6:e27848.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download