Abstract
Purpose
Inadequate empirical therapy for severe infections caused by extended-spectrum β-lactamase-producing Escherichia coli (ESBLEC) is associated with poor outcomes. This study was designed to investigate risk factors for community-onset ESBLEC bacteremia at admission to a tertiary care hospital.
Materials and Methods
A case-control study was performed that included all episodes of ESBLEC bacteremia in the outpatient department or within 48 hours of admission from January 2005 to March 2009. Data on predisposing factors were collected. The molecular epidemiology of ESBLEC clinical isolates was also determined.
Results
Among 25281 blood cultures, 60 episodes of ESBLEC bacteremia were studied, which accounted for 7% of all E. coli bacteremia at admission. Healthcare-associated infection [odds ratio (OR), 8.3; 95% confidence interval (CI), 2.4-28.7; p=0.001], malignancy (OR, 4.6; 95% CI, 1.3-16.3; p=0.018), urinary tract infection (OR, 139.1; 95% CI, 24.6-788.2; p<0.001), hepatobiliary infection (OR, 79.1; 95% CI, 13.5-463.8; p<0.001), third generation cephalosporin usage during preceding 3 months (OR, 16.4; 95% CI, 2.0-131.8; p=0.008), and severe sepsis/septic shock (OR, 73.7; 95% CI, 12.4-438.5; p<0.001) were determined as independent risk factors for community-onset ESBLEC bacteremia. The most common extended-spectrum β-lactamase (ESBL) gene identified was blaCTX-M-15 (n=31) followed by blaCTX-M-14 (n=23).
Conclusion
The most common types of ESBLs in E. coli causing community-onset bacteremia were CTX-M-15 and CTX-M-14 in Korea. By result of decision tree analysis, the empirical use of carbapenems is suggested only for patients with severe sepsis/septic shock, hepatobiliary infection, or healthcare-associated urinary tract infection.
Extended-spectrum β-lactamase-producing Escherichia coli (ESBLEC) is an emerging cause of nosocomial, healthcare-associated, and community-acquired infections worldwide.1 Inadequate empirical antibiotic therapy for infections caused by this microorganism is associated with poor outcomes,2-4 especially in severe infections.5 Considering the increased prevalence of ESBLEC, elucidation of risk factors for ESBLEC bacteremia is critical in terms of empirical treatment of the patients. Although there have been several studies for infections caused by extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae, only a few studies have investigated the risk factors for ESBLEC bacteremia.
The CTX-M enzymes are spreading rapidly and are now the dominant type of ESBL in E. coli in many parts of the world.6 Among CTX-M enzymes, members of the CTX-M-1 and CTX-M-9 clusters have repeatedly been found worldwide including Korea.7
This study was designed to investigate risk factors for community-onset ESBLEC bacteremia at the time of admission to a tertiary care hospital. Decision-tree analysis using the classification and regression tree (CART) algorithm was performed to predict which subgroup of patients who had a blood culture within 48 hours of admission was at increased risk of being infected by ESBLEC bacteremia. The molecular epidemiology of ESBLEC isolates obtained from patients with bacteremia was also determined.
This study was conducted at the Gachon University Gil Medical Center, a 1200 bed tertiary care facility located in Incheon, Republic of Korea. The ESBLEC strains were isolated from the blood cultures of patients from January 2005 through March 2009.
The risk factors for community-onset ESBLEC bacteremia were investigated using a case-control design. A case was defined as an adult (>18 years) with ESBLEC bacteremia that was present in the outpatient department or within 48 hrs of admission to the hospital. Patients with positive blood cultures for ESBLEC, which recovered after 48 hours of admission, were excluded from the study. The first blood isolate per case was studied. Controls were chosen among the patients who had a blood culture performed in the outpatient department or within 48 hrs of admission in the study period if their blood culture did not yield ESBLEC. For each case, three controls were randomly selected. Patients who were ≤18 years old or did not have a blood culture within 48 hrs of admission were excluded from the control group.
Variables analyzed as possible risk factors included age, sex, associated diseases, severity of comorbidity according to the Charlson score,8 healthcare-associated infection, source of bacteremia, invasive procedure such as urinary catheter or tracheostomy during the preceding three months, antimicrobial therapy during preceding three months, presence of severe sepsis or septic shock, and severity of illness as calculated by the Pitt bacteremia score.9
The presence of the following associated diseases was documented: diabetes mellitus, heart failure, chronic pulmonary disease, chronic renal insufficiency, liver cirrhosis, and malignancy. Healthcare-associated infections were classified in accordance with the definition by Friedman, et al.10 with some modifications. Any of the following criteria were considered as healthcare-associated infections: intravenous therapy, wound care, or nursing care received at home 30 days before the bloodstream infection; attendance at a hospital or hemodialysis clinic or receipt of intravenous chemotherapy 30 days before the bloodstream infection; >48-hour hospital admission or performance of invasive procedures such as urinary catheter, endoscopy, and naso-gastric tube 90 days before the bloodstream infection; or residence at a nursing home or long-term care facility. Source of the infection was determined to be the urinary tract, hepatobiliary, gastrointestinal, respiratory, other soft-tissue infection, or primary bloodstream infection.
The study was approved by the Institutional Review Boards of the hospital (GIRBA 2212).
Isolates were identified using a Vitek GNI card (bioMérieux, Marcy l'Etoile, France). Antimicrobial susceptibilities were tested by disk diffusion test on Mueller-Hinton agar (Difco, Cockeysville, MI, USA) and by the agar dilution method according to the interpretative criteria proposed by the Clinical and Laboratory Standards Institute.11 The phenotypic confirmatory test for ESBL and/or AmpC β-lactamase was performed.12 Detection of genes coding for plasmid-borne ESBLs and AmpC β-lactamases was performed by PCR amplification with primers as described previously.7 The templates for PCR amplification in clinical isolates were whole cell lysates, and the PCR products were subjected to direct sequencing. The agar mating method was used to test transferability of oxyimino-cephalosporin resistance determinants using azide-resistant E. coli J53 as a recipient. Transconjugants were selected on Mueller-Hinton agar plates supplemented with 2 µg/mL cefotaxime and 100 µg/mL sodium azide.13
Pulsed-field gel electrophoresis (PFGE) was performed with XbaI restriction enzyme using a CHEF-DRII device (Bio-Rad, Hercules, CA, USA). Tiff format gel images were exported to Molecular Analyst Fingerprinting Software Ver. 3.2 (Bio-Rad) for analysis. Comparisons for E. coli isolates were made by using the band-based dice coefficient. Dendrograms were generated using the unweighted pair group method with arithmetic averages method with 1.0% position tolerance. Multilocus sequence typing (MLST) was performed on ESBLEC isolates using seven conserved housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) following protocols at http://mlst.ucc.ie/mlst/dbs/Ecoli.
Univariate analyses were performed separately for each of the variables. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated for binomial variables. p values were calculated using Fisher's exact test for categorical variables. Variables with a p value of <0.1 in the univariate analysis were candidates for multivariate analysis using a backward elimination method. The area under the receiver operator characteristic (ROC) curve was calculated to evaluate the performance of the models. To split patients into more homogeneous subgroups, a CART analysis was used to build a binary classification tree through recursive partitioning. All tests were 2-tailed, and a p value of <0.05 was considered significant in the multivariable model. STATA software package version 10.0 (StataCorp, College Station, TX, USA) was used to perform the multiple logistic regression, and R 2.4.1 (The R foundation for statistical computing) was used to construct the CART algorithm.
During the study period, 25281 blood cultures were taken from adult patients in the outpatient department or within 48 hours of admission to the Gil Hospital. Among them, 3452 episodes of positive blood cultures (13.7%) were for any organism including 891 E. coli were found, among which 62 were ESBL producers by phenotypic analysis. Since the ESBL gene could not be detected in two isolates, risk factors and microbiological profiles were assessed in 60 patients with ESBLEC bacteremia. Two isolates were excluded from the analysis because they were positive for TEM-1 without SHV-, CTX-M-type ESBL genes.
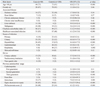
Univariate analysis showed that age >60 years old; liver cirrhosis; malignancy; Charlson comorbidity index; healthcare-associated infection; urinary tract, hepatobiliary, or respiratory tract as the infection source; indwelling urinary catheter; naso-gastric tube; third generation cephalosporin and quinolone usage during preceding three months of admission; septic shock/severe sepsis; and severity of illness as calculated by the Pitt bacteremia score were associated with ESBLEC bacteremia at admission (Table 1). Results of the multivariate logistic regression analysis are presented in Table 2. In the final model, healthcare-associated infection (OR, 8.3; 95% CI, 2.4-28.7; p=0.001), malignancy (OR, 4.6; 95% CI, 1.3-16.3; p=0.018), urinary tract infection (OR, 139.1; 95% CI, 24.6-788.2; p<0.001), hepatobiliary infection (OR, 79.1; 95% CI, 13.5-463.8; p<0.001), third generation cephalosporin usage during preceding 3 months (OR, 16.4; 95% CI, 2.0-131.8; p=0.008) and severe sepsis/septic shock (OR, 73.7; 95% CI, 12.4-438.5; p<0.001) were identified as independent risk factors for ESBLEC bacteremia at admission. The fitness of the final multiple logistic regression model was excellent (area under ROC curve, 0.96; 95% CI, 0.94-0.99). The CART analysis included the six variables significant in the multivariate logistic regression analysis, and identified severe sepsis/septic shock at presentation as the best single discriminator between ESBL bacteremia and no ESBL bacteremia. The next best predictor of ESBL bacteremia in no severe sepsis/septic shock node was urinary tract infection. For the node with patients without severe sepsis/septic shock and no urinary tract infection, hepatobiliary infection provided additional predictive value. For the node with patients without severe sepsis/septic shock, urinary tract infection, or hepatobilary infection, healthcare-associated infection provided additional predictive value (Fig. 1).
All 60 isolates showed positive results in the phenotypic confirmatory test, indicating ESBL production. The most common type of ESBL gene identified was blaCTX-M-15 (n=31) followed by blaCTX-M-14 (n=23). Other types of ESBL gene, such as blaCTX-M-22 (n=2), blaCTX-M-24 (n=1), blaCTX-M-57 (n=1), and blaSHV-12 (n=2) were also identified. Annual percentages of ESBL-producing isolates in community-onset bacteremic E. coli increased from 1% in 2005 to 21% in 2009 (p<0.001). Among ESBLEC isolates harboring blaCTX-M genes, the predominant CTX-M group was the CTX-M-1 group in 2005-2007 and the CTX-M-9 group in 2008-2009 (86% and 57%, respectively; p=0.002) (Table 3). Six ESBL-producing isolates also carried an AmpC gene. The blaCMY-2 gene was identified in three isolates carrying the blaCTX-M-15 gene and one isolate carrying the blaCTX-M-14 gene. Both isolates carrying the blaSHV-12 gene also carried the blaDHA-1 gene. Despite repeated attempts, transconjugants were obtained from only 21 of 60 isolates.
After all of the 60 isolates were subjected to MLST analysis, the most commonly identified sequence type (ST) was ST131 (n=11) followed by ST648 (n=6), ST38 (n=4), ST405 (n=4), ST69 (n=3), ST95 (n=3), and ST410 (n=3). Other STs, including ST10, ST44, ST46, ST112, ST354, ST393, ST457, ST617, ST773, ST939, ST964, ST1011, ST1177, and ST2037, were found in one or two isolates. The ST2037 (85-88-78-37-59-58-62) was newly identified in this study. The four of 11 E. coli isolates of ST131 harbored the blaCTX-M-15 gene. The remaining seven isolates of ST131 carried blaCTX-M-14 (n=6) or blaSHV-12 (n=1). Banding patterns of XbaI-digested DNA showed less than 80% genetic similarity for most of the CTX-M-type- or SHV-12-producing isolates, which were considered to be unrelated (Figs. 2 and 3).
This study was designed to identify the best predictors and molecular epidemiology of community-onset ESBLEC bloodstream infection. Because ESBL-producing bacteria are often resistant to various antimicrobials including fluoroquinolones and oxyimino-cephalosporins, the presence of these enzymes complicates the selection of empirical antibiotics while the results of cultures and antimicrobial susceptibility profiles are awaited. Several studies have addressed the impact of inadequate empirical antimicrobial treatment in patients with infections caused by ESBL producers.3,14 A delay or failure in initiating adequate antimicrobial therapy was associated with increased morbidity and mortality in severe infections.5,15 Moreover, in septic patients, the importance of appropriate empirical therapy has also been emphasized.16-18 For effective empirical treatment, risk factor analysis for any bloodstream infection by ESBL producers has important clinical implications. Although there have been numerous reports of infections or colonization with ESBL-producing organisms in recent years, only five studies have been conducted to analyze the risk factors of ESBLEC bloodstream infections.19-23 Although two studies have investigated the risk factors for community-onset bloodstream infections of ESBLEC,21,22 as risk factors may differ according to geographic variation and patient population, empirical antibiotic choices should be individualized.
In the present study, we selected control patients from all adult patients who had a blood culture in the outpatient department or within 48 hours of admission to Gil Hospital to investigate the risk factors of ESBLEC bacteremia among patients who were septic; therefore blood cultures were taken at presentation. Although some of the documented risk factors in this study might be those of a susceptible organism (non-ESBL producing E. coli),24 we believe that our results certainly give clinical clues as to what subpopulation of the patients need empirical therapy for ESBLEC bacteremia. If patients infected with the antimicrobial susceptible organism were used as control patients, these 'susceptible control patients' (i.e., patients with non-ESBL producing E. coli bacteremia) would not be representative of the 'source population' for antimicrobial resistant organisms, and this may lead to an overestimation of the association between antimicrobial exposure and cases.24-26
The CART analysis is a statistical method based on a recursive partitioning analysis. Unlike multivariate logistic regression, by which the odds ratio produced can be difficult to translate, CART is well suited clinical decisions on rules and produces decision trees that are simple to interpret. CART has been successfully used to assist in the diagnosis of various clinical conditions including infections27,28 and neurological,29 oncological,29,30 and cardiac disorders.31 According to our results, the empirical use of carbapenems is suggested for patients with severe sepsis/septic shock, hepatobiliary infection, or healthcare-associated urinary tract infection. Before the present risk factors might be applied to clinical decisions for empirical therapy, the limitations for our study must be considered. This study was performed with a relatively small number of patients in a single center. In addition, documented risk factors in our study were based on data that is several years old. Clinicians should take into account that the epidemiology of Enterobacteriaceae changes rapidly, which was underlined by the major change in ESBLEC prevalence in this study population. Moreover, increasing carbapenem resistance, mediated by porin loss or carbapenemases, should be considered32 because the presence of carbapenem-resistant Enterobacteriaceae has been reported worldwide including the Republic of Korea.33
The predominance of CTX-M enzymes and clonally unrelated isolates are consistent with the fact that ESBLEC is a true community pathogen. Although healthcare-associated infection was associated with an increased risk of ESBLEC bacteremia in our study, it could be a surrogate marker because ESBLEC colonization is mainly a problem in the community.1,34,35 Although CTX-M-9 group enzymes are the most prevalent ESBLs in Spain, China, and Taiwan, currently, the most prevalent CTX-M enzyme worldwide is CTX-M-15, which has been reported in Europe, Asia, Africa, North America, South America, and Australia.36 The predominance of CTX-M-15 and CTX-M-14 is in agreement with recent reports in Korea.37-39 Moreover, the presence of the CTX-M-9 group is increasing in Korea. In the present study, the most predominant CTX-M group was the CTX-M-1 group in 2005-2007 and the CTX-M-9 group in 2008-2009 among all ESBLEC isolates having blaCTX-M genes (86% and 57%, respectively, p=0.002). The rate of ESBL production among E. coli from the blood of patients with community-onset bacteremia was 7% and increased 21 times from 2005 to 2009. This change may be attributable to the dissemination of CTX-M enzymes since the blaCTX-M genes were detected in 58 (97%) of 60 ESBLEC isolates. Further studies are required to obtain more information on the prevalence of ESBL producers and changes to the molecular epidemiology of CTX-M enzymes in the Republic of Korea.
The E. coli O25:H4-ST131 has been recognized as an emerging intercontinental clonal group expressing CTX-M-type ESBL. Most previous studies from Europe and North America reported E. coli O25:H4-ST131 strains were CTX-M-15 producers.40-42 Our analysis of 60 community-onset ESBLEC determined that E. coli ST131 was the most prevalent (18%) clonal group. The E. coli ST131 isolates carried not only the blaCTX-M-15 but also blaCTX-M-14 or blaSHV-12. Although E. coli ST131 was the most common cause of community-onset ESBLEC infection, clonal expansion of strains carrying blaCTX-M-15 was not the main reason for the rapid spread in the community. This hypothesis is supported by the fact that these isolates show low level of similarity in PFGE analysis. Despite that many different types of STs among ESBLEC and the isolates showed a low-level similarity by PFGE, certain ESBL genes, such as blaCTX-M-14 or blaCTX-M-15, were predominantly found among the isolates. This may suggest that the plasmid or conjugative transposons harboring these antibiotic resistance genes were transferred horizontally among E. coli strains. The multi-clonality of ESBLEC, shown in our study, comes not only from the type of ESBL genes but also from the whole genome including virulence and housekeeping genes, so a multi-clonal outbreak of ESBLEC does not always correlate with the diversity of ESBL genes. This means that the predominance of certain type of ESBL genes may not result in the mono-clonal spreading of ESBLEC.
Our results showed that physicians who care for patients with these risk factors should consider ESBLEC as the causative organism of community-onset bacteremia. The most common types of ESBLs in E. coli causing community-onset bacteremia were CTX-M-15 and CTX-M-14 in the Republic of Korea, which might have transferred horizontally.
Figures and Tables
Fig. 1
Classification and regression trees analysis for predicting extended-spectrum β-lactamase (ESBL)-producing Escherichia coli bacteremia among adult patients who had a blood culture within 48 hours of admission to tertiary care hospital.

Fig. 2
Dendrogram based on XbaI-macrorestriction patterns of E. coli isolates producing CTX-M-1-type ESBLs. The dashed line indicates 80% similarity. E. coli isolates exhibiting similarities of <80% were considered unrelated. *XbaI-macrorestriction analysis yielded no DNA banding patterns due to the degeneration of the genomic DNA during preparation of the agarose plugs. ESBL, extended-spectrum β-lactamase.

Fig. 3
Dendrogram based on XbaI-macrorestriction patterns of E. coli isolates producing CTX-M-9-type and SHV ESBLs. The dashed line indicates 80% similarity. E. coli isolates exhibiting similarities of <80% were considered unrelated. *XbaI-macrorestriction analysis yielded no DNA banding patterns due to the degeneration of the genomic DNA during preparation of the agarose plugs. ESBL, extended-spectrum β-lactamase

Table 1
Univariate Analysis of Risk Factors for Community-Onset ESBL-Producing Escherichia coli Bacteremia
ACKNOWLEDGEMENTS
This study was supported by a research grant from the Department of Internal Medicine, Gachon University (2009).
References
1. Pitout JD, Nordmann P, Laupland KB, Poirel L. Emergence of Enterobacteriaceae producing extended-spectrum beta-lactamases (ESBLs) in the community. J Antimicrob Chemother. 2005; 56:52–59.

2. Lautenbach E, Patel JB, Bilker WB, Edelstein PH, Fishman NO. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Clin Infect Dis. 2001; 32:1162–1171.

3. Tumbarello M, Sanguinetti M, Montuori E, Trecarichi EM, Posteraro B, Fiori B, et al. Predictors of mortality in patients with bloodstream infections caused by extended-spectrum-beta-lactamase-producing Enterobacteriaceae: importance of inadequate initial antimicrobial treatment. Antimicrob Agents Chemother. 2007; 51:1987–1994.

4. Rodríguez-Baño J, Navarro MD, Romero L, Muniain MA, de Cueto M, Ríos MJ, et al. Bacteremia due to extended-spectrum beta-lactamase-producing Escherichia coli in the CTX-M era: a new clinical challenge. Clin Infect Dis. 2006; 43:1407–1414.
5. Hyle EP, Lipworth AD, Zaoutis TE, Nachamkin I, Bilker WB, Lautenbach E. Impact of inadequate initial antimicrobial therapy on mortality in infections due to extended-spectrum beta-lactamase-producing enterobacteriaceae: variability by site of infection. Arch Intern Med. 2005; 165:1375–1380.

6. Rossolini GM, D'Andrea MM, Mugnaioli C. The spread of CTX-M-type extended-spectrum beta-lactamases. Clin Microbiol Infect. 2008; 14:Suppl 1. 33–41.
7. Ryoo NH, Kim EC, Hong SG, Park YJ, Lee K, Bae IK, et al. Dissemination of SHV-12 and CTX-M-type extended-spectrum beta-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae and emergence of GES-3 in Korea. J Antimicrob Chemother. 2005; 56:698–702.

8. Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987; 40:373–383.

9. Paterson DL, Ko WC, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, et al. International prospective study of Klebsiella pneumoniae bacteremia: implications of extended-spectrum beta-lactamase production in nosocomial Infections. Ann Intern Med. 2004; 140:26–32.

10. Friedman ND, Kaye KS, Stout JE, McGarry SA, Trivette SL, Briggs JP, et al. Health care--associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med. 2002; 137:791–797.

11. Cockerill FR. Performance Standards for Antimicrobial Susceptibility Testing; Twentieth Informational Supplement. Wayne, PA: CLSI;2010.
12. Song W, Jeong SH, Kim JS, Kim HS, Shin DH, Roh KH, et al. Use of boronic acid disk methods to detect the combined expression of plasmid-mediated AmpC beta-lactamases and extended-spectrum beta-lactamases in clinical isolates of Klebsiella spp., Salmonella spp., and Proteus mirabilis. Diagn Microbiol Infect Dis. 2007; 57:315–318.

13. Jacoby GA, Han P. Detection of extended-spectrum beta-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol. 1996; 34:908–911.

14. Anderson DJ, Engemann JJ, Harrell LJ, Carmeli Y, Reller LB, Kaye KS. Predictors of mortality in patients with bloodstream infection due to ceftazidime-resistant Klebsiella pneumoniae. Antimicrob Agents Chemother. 2006; 50:1715–1720.

15. Peña C, Gudiol C, Calatayud L, Tubau F, Domínguez MA, Pujol M, et al. Infections due to Escherichia coli producing extended-spectrum beta-lactamase among hospitalised patients: factors influencing mortality. J Hosp Infect. 2008; 68:116–122.

16. Harbarth S, Garbino J, Pugin J, Romand JA, Lew D, Pittet D. Inappropriate initial antimicrobial therapy and its effect on survival in a clinical trial of immunomodulating therapy for severe sepsis. Am J Med. 2003; 115:529–535.

17. Kang CI, Kim SH, Park WB, Lee KD, Kim HB, Kim EC, et al. Bloodstream infections caused by antibiotic-resistant gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob Agents Chemother. 2005; 49:760–766.

18. Kumar A, Ellis P, Arabi Y, Roberts D, Light B, Parrillo JE, et al. Initiation of inappropriate antimicrobial therapy results in a fivefold reduction of survival in human septic shock. Chest. 2009; 136:1237–1248.

19. Rodríguez-Baño J, Navarro MD, Romero L, Muniain MA, Cueto Md, Gálvez J, et al. Risk-factors for emerging bloodstream infections caused by extended-spectrum beta-lactamase-producing Escherichia coli. Clin Microbiol Infect. 2008; 14:180–183.

20. Ho PL, Chan WM, Tsang KW, Wong SS, Young K. Bacteremia caused by Escherichia coli producing extended-spectrum beta-lactamase: a case-control study of risk factors and outcomes. Scand J Infect Dis. 2002; 34:567–573.

21. Rodríguez-Baño J, Picón E, Gijón P, Hernández JR, Ruíz M, Peña C, et al. Community-onset bacteremia due to extended-spectrum beta-lactamase-producing Escherichia coli: risk factors and prognosis. Clin Infect Dis. 2010; 50:40–48.

22. Kang CI, Song JH, Chung DR, Peck KR, Ko KS, Yeom JS, et al. Risk factors and treatment outcomes of community-onset bacteraemia caused by extended-spectrum beta-lactamase-producing Escherichia coli. Int J Antimicrob Agents. 2010; 36:284–287.

23. Gudiol C, Calatayud L, Garcia-Vidal C, Lora-Tamayo J, Cisnal M, Duarte R, et al. Bacteraemia due to extended-spectrum beta-lactamase-producing Escherichia coli (ESBL-EC) in cancer patients: clinical features, risk factors, molecular epidemiology and outcome. J Antimicrob Chemother. 2010; 65:333–341.

24. Kaye KS, Harris AD, Samore M, Carmeli Y. The case-case-control study design: addressing the limitations of risk factor studies for antimicrobial resistance. Infect Control Hosp Epidemiol. 2005; 26:346–351.

25. Harris AD, Karchmer TB, Carmeli Y, Samore MH. Methodological principles of case-control studies that analyzed risk factors for antibiotic resistance: a systematic review. Clin Infect Dis. 2001; 32:1055–1061.

26. Harris AD, Samore MH, Carmeli Y. Control group selection is an important but neglected issue in studies of antibiotic resistance. Ann Intern Med. 2000; 133:159.

27. Gerald LB, Tang S, Bruce F, Redden D, Kimerling ME, Brook N, et al. A decision tree for tuberculosis contact investigation. Am J Respir Crit Care Med. 2002; 166:1122–1127.

28. Kammerer JS, McNabb SJ, Becerra JE, Rosenblum L, Shang N, Iademarco MF, et al. Tuberculosis transmission in nontraditional settings: a decision-tree approach. Am J Prev Med. 2005; 28:201–207.
29. Temkin NR, Holubkov R, Machamer JE, Winn HR, Dikmen SS. Classification and regression trees (CART) for prediction of function at 1 year following head trauma. J Neurosurg. 1995; 82:764–771.

30. Kvasnicka HM, Thiele J, Schmitt-Graeff A, Diehl V, Zankovich R, Niederle N, et al. Bone marrow features improve prognostic efficiency in multivariate risk classification of chronic-phase Ph(1+) chronic myelogenous leukemia: a multicenter trial. J Clin Oncol. 2001; 19:2994–3009.

31. Fonarow GC, Adams KF Jr, Abraham WT, Yancy CW, Boscardin WJ. ADHERE Scientific Advisory Committee, Study Group, and Investigators. Risk stratification for in-hospital mortality in acutely decompensated heart failure: classification and regression tree analysis. JAMA. 2005; 293:572–580.

32. Livermore DM. Current epidemiology and growing resistance of gram-negative pathogens. Korean J Intern Med. 2012; 27:128–142.

33. Kim SY, Shin J, Shin SY, Ko KS. Characteristics of carbapenem-resistant Enterobacteriaceae isolates from Korea. Diagn Microbiol Infect Dis. 2013; 76:486–490.

34. Rodríguez-Baño J, Alcalá JC, Cisneros JM, Grill F, Oliver A, Horcajada JP, et al. Community infections caused by extended-spectrum beta-lactamase-producing Escherichia coli. Arch Intern Med. 2008; 168:1897–1902.

35. Pitout JD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008; 8:159–166.

36. Peirano G, Pitout JD. Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int J Antimicrob Agents. 2010; 35:316–321.

37. Song W, Lee H, Lee K, Jeong SH, Bae IK, Kim JS, et al. CTX-M-14 and CTX-M-15 enzymes are the dominant type of extended-spectrum beta-lactamase in clinical isolates of Escherichia coli from Korea. J Med Microbiol. 2009; 58(Pt 2):261–266.

38. Kang CI, Wi YM, Lee MY, Ko KS, Chung DR, Peck KR, et al. Epidemiology and risk factors of community onset infections caused by extended-spectrum β-lactamase-producing Escherichia coli strains. J Clin Microbiol. 2012; 50:312–317.

39. Park SH, Byun JH, Choi SM, Lee DG, Kim SH, Kwon JC, et al. Molecular epidemiology of extended-spectrum β-lactamase-producing Escherichia coli in the community and hospital in Korea: emergence of ST131 producing CTX-M-15. BMC Infect Dis. 2012; 12:149.

40. Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis. 2010; 51:286–294.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download