Abstract
Purpose
Candida albicans is an opportunistic pathogen that is commonly found in human microflora. Biofilm formation (BF) is known as a major virulence factor of C. albicans. The aim of this study was to examine the influence of bacterial presence on biofilm formation of C. albicans.
Materials and Methods
The BF of Candida was investigated when it was co-cultured with C. albicans (C. albicans 53, a yeast with a low BF ability, and C. albicans 163, a yeast with high BF ability) and bacteria. BF was assessed with XTT reduction assay. A scanning electron microscope was used to determine the structure of the biofilm, and real-time reverse transcriptase polymerase chain reaction was used to amplify and quantify hyphae-associated genes.
Results
Co-culturing with two different types of bacteria increased the BF value. Co-culturing with C. albicans 53 and 163 also increased the BF value compared to the value that was obtained when the C. albicans was cultured individually. However, co-culturing with bacteria decreased the BF value of C. albicans, and the BF of C. albicans 163 was markedly inhibited. The expression of adherence and morphology transition related genes were significantly inhibited by co-culturing with live bacteria.
Most microorganisms in their natural form, as driven by the environmental conditions of their habitats, stay as free-floating cells or exist in a planktonic state or as biofilms that adhere to and grow on extracellular surfaces.1 When nutrients are depleted, the response is transformation into the state of biofilm.2 In addition, a number of physical, biological and chemical processes are involved in the formation of biofilm.
The biological significance of the biofilm in C. albicans is that it protects the bacteria by delaying its physical destruction and acts as a protective barrier against host immune substances and the penetration of anti-fungal agents.3 The formation of biofilm in C. albicans occurs in three stages. The first stage is adherence to a suitable temperament basal layer; this is initiated after the start of incubation. This initial adherence stage is regulated by non-specific factors, such as hydrophobic or electrostatic interaction, and specific factors through specific receptors like fibrinogen and fibronectin.4,5 The second stage is the process in which the adhered C. albicans continuously multiply and the yeast cells transform into hyphae that then form three-dimensional structures. The three-dimensional structures of biofilm are generally comprised of yeast, pseudohyphae, and hyphae.6,7 The last maturation stage is the process of quantitative increase of extracellular substances; the mature biofilm enables the Candida yeast to fix biofilm on-to the extracellular surface, and the hyphae form a cross-sectional structure with structural frames.8 As described, the dimorphism of C. albicans serves as a major factor that influences the formation of biofilm.9,10
In the human body, a variety of bacteria and fungi interact and coexist regardless of whether the host is in a healthy or diseased state.11-13 Most interactions among complex strains comprise a colony by one microorganism, and the colony is either commensal in which the existence of the other microorganisms is beneficial or antagonistic in which microorganisms with differing microbial products inhibit, wound, or kill the growth of other microorganisms.2,14,15 Recognition of other populations by bacteria or fungi can be explained by Quorum Sensing, and bacteria are known to control the formation of microfilm with respect to the density and growth rates of the other surrounding populations.16-19
The C. albicans infection that appears on inserted medical devices is caused by the formation of C. albicans microfilm. The fact that biofilm is observed in many cases when Candida and other various bacteria are mixed together warrants attention. Previous studies on biofilm development have been focused on the interactions of bacterial species and have not included the study of interactions between bacteria and fungi. In this study, attempts were made to determine how the existence of bacteria affected the formation of the microfilm, by first identifying the biofilm that is formed when bacteria and fungi are incubated individually. The second aim was to identify the biofilm that is formed when bacteria-and-bacteria or fungi-and-fungi incubations occur. The third aim was to identify the biofilm that is formed by complex strains of co-cultured bacteria and fungi.
Clinical isolates of Candida albicans were obtained; one commensal strain was isolated from the oral cavity of a healthy volunteer and the other was isolated from the blood of a patient. Escherichia coli, Pseudomonas aeruginosa, and Proteus vulgaris were isolated from urine of a patient. In addition, Staphylococcus aureus, Streptococcus pyogenes, and Streptococcus salivarius were isolated from blood of a patient. The identity of all strains was confirmed using the API 20E identification systems (BioMerieux, Marcy I'Etoile, France) for P. aeruginosa, P. vulgaris and E. coli, the API 20STREP identification system for streptococci and the API STAPH identification system for staphylococci.
Prior to each experiment, C. albicans isolates were cultured at 30℃ for 18 h on Sabouraud's dextrose agar (Difco™, Becton Dickinson, Spark, MD, USA), and one colony of bacteria was inoculated into yeast nitrogen base (Difco™) medium supplemented with 50 mM glucose. The two streptococcal species used were S. pyogenes and S. salivarius, which were first sub-cultured at 37℃ for two days on blood agar. One colony of bacteria was inoculated into brain heart infusion (Difco™) medium and incubated at 37℃ in a 5% CO2 incubator for two days. E. coli, P. aeruginosa, P. vulgaris, and S. aureus were first subcultured at 37℃ for 18 h on tryptic soy agar. One colony of each bacterium was then inoculated into tryptic soy broth (TSB, Difco™) and incubated at 37℃ for 18 h.
Biofilm formation was quantified using the method developed by Ramage, et al.14 Biofilms were allowed to form on commercially available pre-sterilized, polystyrene, flat-bottom 96-well microtiter plates (Costar, Cambridge, MA, USA). C. albicans (OD. 0.2) was cultured alone. C. albicans (OD600=0.1) and each strain of bacteria (OD600=0.1) were prepared and transferred into selected wells of a microtiter plate. The plate was incubated for 90 min at 37℃ in an orbital shaker at 75 rpm. After the initial adhesion phase, the cell suspensions were aspirated, and each well was washed twice with phosphate-buffered saline (PBS) to remove loosely adherent cells. A volume of 200 µL of medium was added to each well, and the plate was then incubated for another 72 h. After biofilm formation, the medium was aspirated, and non-adherent cells were removed by thoroughly washing the biofilm three times with PBS. A quantitative measure of biofilm formation was calculated using XTT [2,3-bis(2-methyoxy-4-nitro-5-sulfo-phenyl)-2H-tetrazolium-5-carboxanilide]-reduction assay. A 200 µL aliquot of XTT (1 mg/mL, Sigma, St. Louis, MO, USA) and menadione (0.4 mM, Sigma) solution was then added to each well containing the prewashed biofilm and the control well. The plates were then incubated in the dark for up to 3 h at 37℃. A colorimetric change resulting from XTT reduction was measured using a microtiter plate reader (Emax, Molecular Devices, Sunnyvale, CA, USA) at 490 nm.
We developed biofilms from single species as well as candidal biofilms that were co-cultured with bacteria on polystyrene coverslips as described. The coverslips were washed twice with PBS and placed in PBS with a fixative of 2.5% glutaraldehyde (Sigma) for 20 h. After, they were washed for 5 min in PBS and then placed in 1% osmium tetroxide for 30 min. After a series of alcohol washes, a final drying step was performed using the critical point drying method. Biofilms were then mounted and gold coated. Samples were imaged with a scanning electron microscope (TM-1000, Hitachi, Tokyo, Japan) in high-vacuum mode at 15 kV.
RNA was isolated from C. albicans 163 cells using the MasterPure Yeast RNA Extraction kit (Epicentre Biotechnologies, Post Rd, Madson, WI, USA). RNA was treated with amplification grade DNase I (Epicentre) and used for cDNA synthesis with random hexamer primer (Invitrogen Life Technologies, Carlsbad, CA, USA) using Superscript II reversetranscriptase reagents (Invitrogen Life Technologies). Each reaction contained 1 µg of total RNA, 1 µL hexamer, 50 µM and 1 µL dNTP 10 mM in a final volume of 10 µL. Reactions were incubated at 65℃ for 5 min and cooled on ice. To each reaction tube, 10 µL of the following mixture was added: 4 µL of 5 X First-Strand Buffer, 2 µL MgCl2 10 mM, 2 µL DTT 0.1 M, 1.4 µL RNAse inhibitor and 1 µL Superscript II. Reactions were incubated at 42℃ for 50 min and then at 70℃ for 15 min.
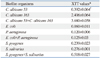
Real-time polymerase chain reaction (PCR) contained 10 µL of Power SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA) as well as a forward and reverse primer (1 µL of each) (Table 1)17,18 and sterile water to make a final volume of 20 µL. The PCR was run on MicroAmp® Optical 384-well reaction plates in an ABI 7900 Real-Time PCR system (Applied Biosystems). Real-time PCR reactions were performed at 95℃ for 5 min, followed by 40 cycles of 15 sec at 95℃ and 1 min at 60℃. Dissociation curves were analyzed for all reactions to verify single peaks/products. Expression levels were analyzed using ABI 7900 System SDS software (Applied Biosystems). Real-time PCR data were normalized with the geometric mean of two reference genes. ACT1 and PMA1 were used for this purpose.
All experiments were performed in triplicate on three different occasions. All data were expressed as mean values with corresponding standard deviations. Student's t-tests and Mann-Whitney U tests were used to compare the differences between Candida only and Candida co-cultured with bacteria and a p-value of <0.05 was considered statistically significant.
The biofilm value generated when each type of bacteria was incubated separately was 0.048±0.026 for E. coli, 0.081±0.030 for P. aeruginosa, 0.072±0.008 for P. vulgaris, 0.155±0.011 for S. aureus, 0.239±0.008 for S. pyogenes, and 0.276±0.014 for S. salivarius (Fig. 1). These values were confirmed, and we hypothesized that Gram-positive bacteria formed a microfilm more often than that of Gram-negative bacteria.
C. albicans 53 is a strain isolated from normal oral flora, and the biofilm formation OD value was 0.392±0.064 when it was incubated alone. C. albicans 163 is a strain isolated from blood, and the biofilm formation OD value was 2.406±0.064 when it was incubated alone. When the two aforementioned C. albicans strains were co-incubated, the biofilm value increased sharply to 3.680±0.058. When the Gram-negative bacteria E. coli and P. aeruginosa were incubated separately, the biofilm value was 0.06±0.011 and 0.120±0.006, respectively, and when they were co-incubated, the biofilm value was 0.125±0.03. When the Gram-positive bacteria S. pyogenes and S. salivarius were incubated separately, the value was 0.239±0.023 and 0.276±0.001, respectively, and when they were co-incubated, the value was 0.518±0.027 (Table 2). In each of these three cases, the amount of biofilm formed when two bacteria were incubated together was equal to the sum of the two biofilm values when the bacteria were incubated separately. However, in the case of fungi, the value was much higher when incubated together than the sum of these values when they were incubated separately. In other words, bacteria-bacteria incubation has an additive effect, whereas, fungi-fungi incubation has a synergistic effect.
The formations of biofilm for C. albicans incubated alone and when it was incubated along with six kinds of bacteria were compared. The biofilm value when C. albicans 53 was incubated alone was 0.392±0.064, and when it was incubated with E. coli, the biofilm value was 0.177±0.040. When the C. albicans was incubated with P. aeruginosa, P. vulgaris, S. aureus, S. pyogenes, S. salivarius, the biofilm values were 0.120±0.050, 0.079±0.006, 0.162±0.015, 0.298±0.026, and 0.198±0.053, respectively (Fig. 2A). C. albicans 163, which has a high biofilm formation when incubated alone, had a biofilm formation as measured using OD of 2.406±0.064. When incubated with each of the six different types of bacteria separately, the value of the biofilm when incubated with E. coli was 0.182±0.033, with P. aeruginosa 0.197±0.029, with P. vulgaris 0.143±0.051, with S. aureus 0.237±0.044, with S. pyogenes 0.349±0.024, and with S. salivarius 0.157±0.032 (Fig. 2B). Both the Candida with low biofilm formation and the C. albicans with high biofilm formation had lower biofilm formation when incubated with other types of bacteria than when incubated alone. The strains with high biofilm formation also showed pronouncedly low biofilm formation in the presence of bacteria.
To assess the effect of dead bacteria on the biofilm formation of C. albicans, C. albicans were incubated with each type of tested bacteria that had been heat-treated for 30 min at 100℃. The value of the biofilm formation when the C. albicans 53 strain was incubated with heat-treated E. coli was 0.105±0.013. This value was 0.118±0.019 when this strain was incubated with heat-treated P. aeruginosa, 0.153±0.007 when incubated with heat-treated P. vulgaris, 0.215±0.028 when incubated with heat-treated S. aureus, 0.085±0.012 when incubated with heat-treated S. pyogenes and 0.111±0.017 when incubated with heat-treated S. salivarius (Fig. 3A). The value of the biofilm formation when C. albicans 163 and heat-treated E. coli were co-incubated was 0.519±0.067. This value was 0.631±0.100 when this strain of C. albicans was incubated with heat-treated P. aeruginosa, 0.643±0.204 when incubated with heat-treated P. vulgaris, 0.637±0.079 when incubated with heat-treated S. aureus, 0.512±0.037 when incubated with heat-treated S. pyogenes, and 0.534±0.033 when incubated with heat-treated S. salivarius (Fig. 3B). Thus, our findings were able to verify that the biofilm formation of C. albicans is reduced by 50% to 80% when incubated with dead bacteria.
Changes were observed in the biofilm formation of bacteria incubated alone and that co-cultured with C. albicans heat-treated at 100℃ for 30 min in order to provide a basal layer. When heat-treated C. albicans was incubated with E. coli, the value of the biofilm was 0.033±0.011, 0.100±0.030 when incubated with P. aeruginosa, 0.063±0.010 when incubated with P. vulgaris, 0.146±0.012 when incubated with S. aureus, 0.249±0.005 when incubated with S. pyogenes and 0.189±0.012 when incubated with S. salivarius. The cases in which the biofilm formation of the suspension with the dead C. albicans were similar to the suspensions containing only bacteria demonstrates that dead bacteria affected C. albicans biofilm formation, but that dead C. albicans did not affect bacteria biofilm formation (Fig. 4).
When biofilm is formed by adhering to the extracellular surface of a microorganism, it grows from a single-layer structure to a multi-layer three-dimensional structure. Structural differences exist depending on whether the formed biofilm develops in C. albicans 163 strain and whether bacteria is incubated individually or together, and this difference has been confirmed by scanning electron microscopy. The biofilm of C. albicans when incubated alone appeared to be high in density and was a multi-layer solid (Fig. 5A). The biofilm formed when C. albicans and bacteria were incubated together showed that the bacteria were attached to hyphae of the C. albicans and inserted between the C. albicans cells (Fig. 5B). The thickness of the biofilm in these cases also appeared to be thin and low in density. This also confirmed that biofilm formation was low in cases where C. albicans were incubated with heat-treated bacteria (Fig. 5C).
We performed quantitative reverse transcriptase PCR to assess the expression of hyphae-specific genes on during biofilm development of C. albicans. Over time, the expression of hyphae-specific genes (Als3, Ece1, Hwp1, Sap5) were strongly enhanced (Fig. 6). To clarify the inhibitory effect of mixed cultures on biofilm formation, we analyzed changes in the gene expression levels of C. albicans in biofilm co-cultured with indicated bacteria. The transcription levels of hyphae-specific genes were remarkably reduced by C. albicans co-cultured with bacteria during the early stage of biofilm development (Fig. 7). The transcription levels of hyphae-specific genes were further reduced by C. albicans co-cultured with gram-negative bacteria (P. aeruginosa, P. vulgaris and E. coli) than with gram-positive bacteria (S. aureus, S. pyogenes, and S. salivarius). This was the most significant reduction of mRNA expression of hyphae-specific genes when C. albicans cultured with P. vulgaris.
Previous studies on biofilm have been based on only a single type of microorganism and, in particular, have focused mostly on bacteria that are associated with pathogenicity.19 However, biofilms that are discovered outside laboratories are not only produced by various bacteria, but also are comprised of forms that include eukaryotes. Nearly 60% to 70% of recent hospital infections have been associated with medical devices that are inserted into the body, and it has been verified that bacteria and fungi interact together to form such biofilms.20 C. albicans is the fungus that is most often associated with biofilm formation on medical devices, and the resulting C. albicans is associated with a high fatality rate. The first stage of C. albicans biofilm formation is the adherence to a surface, after which, along with bacterial proliferation, the biofilm is mixed with various extracellular polymers to form colonies with a solid structure. In order for C. albicans to adhere to a surface, non-specific factors, such as cell-surface hydrophobicity and electrostatic force, and specific factors, such as adhesins and receptors, are required.4,5
In situations where C. albicans and bacteria are mixed and incubated together, the architecture of the biofilm and the consequential biological functions have not been accurately identified because the interactions among the strains created from a mixed-incubation environment are not well understood.
In this study, when two different types of bacteria were incubated together, the amount of biofilm that was formed showed an additive effect in that it was the sum of the biofilms formed by the two bacteria incubated separately. However, when two different C. albicans strains were incubated together, the resulting amount of biofilm was much greater than the sum of the biofilms when the two strains were incubated separately, and this may be attributable to the two types of C. albicans fungi that provide structural assistance to the formation of biofilm when in culture.
The biofilm formation of C. albicans was reduced when C. albicans suspension was incubated with each of the six different types of bacteria compared to the biofilm formed when the C. albicans was incubated alone. The reduction in biofilm formation was more pronounced in the strains with high biofilm formation. When P. aeruginosa was incubated with C. albicans, it inhibited the biofilm formation of C. albicans, consistent with the known reports of the ability of P. aeruginosa to inhibit the biofilm formation of C. albicans by suppressing the hyphae formation of C. albicans and eventually inducing death by secreting neurotransmitters.21,22 These results were equivalent to the results of the studies about E. coli, a bacteria that is present in the intestines, being able to suppress biofilm formation of C. albicans by strongly inhibiting it from adhering to an extracellular surface.23
Biofilm formation of C. albicans was also reduced when incubated with heat-treated bacteria. The inhibiting action of bacteria was thought to induce not only a metabolite that synthesizes with the bacteria during the phase of multiplication and growth, but also structural changes in the formation of the biofilm, which thereby reduce the biofilm formation of C. albicans. Although the biofilm of C. albicans is inhibited by bacteria, the mere presence of C. albicans does not have an effect on bacterial biofilm formation. C. albicans biofilm is formed through many steps and has a complex structure, while bacterial biofilms have more simplistic structures.
In a scanning electron microscope, it was shown that the C. albicans biofilm was made up of several layers, was highly dense, and was developed evenly throughout the surface, while the E. coli biofilm consists of a single layer, was of low density and was concentrated in one location. When the complex strains of C. albicans and E. coli were co-cultured together, the magnified image of the biofilm, showed that the E. coli strains were located between the C. albicans, and that the density of the biofilm was decreased compared with Candida alone.
Adhesins, such as Hwp1 and Als3, were required on the surface of yeast-form cells for biofilm formation and appear to directly interact with each other during the biofilm formation process.24,25 Moreover, the expression of Ece1 correlated with Candida hyphal elongation,26 and the Sap5 gene was expressed in biofilm associated with mucosal surfaces.27 Using real-time PCR, we accessed the expression of adherence and morphology transition related genes such as Sap5, Als3, Ece1, and Hwp1. These genes were up-regulated as biofilm formation progressed. Interestingly, expression of the Candida genes were significantly inhibited by co-culturing with live bacteria, but were not changed by co-culturing with dead bacteria (data not shown).
In this study, gram-negative bacteria (P. aeruginosa, P. vulgaris and E. coli) reduced C. ablicans biofilm more than that of gram-positive bacteria (S. aureus, S. pyogenes, and S. salivarius). Interestingly, gram-negative bacteria are motile while gram-positive bacteria are non-motile. In particular, P. vulgaris, with the highest motile activity,28,29 remarkably inhibited the biofilm formation of C. albicans. Moreover, these results correspond to the mRNA expression levels of hyphae-specific genes. This suggests that gram-negative cell wall constitutions and/or motile activity of bacteria may inhibit biofilm formation of C. albicans.
In conclusion, the growth of C. albicans was inhibited by bacteria (data not shown) and the formation of Candida biofilms was also inhibited. Additionally, these results were obtained with not only true of live bacteria, but also of dead bacteria. Bacterial mass may inhibit C. albicans biofilm formation as structural discordance. Bacterial growth may influence the down-regulation of hyphal transition-associated gene expression. Future studies are needed to identify the differences in biofilm formation in cases of mixed-incubation by classifying the bacteria into particular types and should enable the bacteria to coexist with the C. albicans fungus. Future studies should also aim to identify the factors that affect biofilm formation due to the interaction between the C. albicans fungus and different types of bacteria.
Figures and Tables
Fig. 1
Biofilm formation was monitored for the different types of bacteria. Suspensions of each type of bacteria (OD. 0.2) were added to wells in a 96-well microtiter plate. The plate was incubated for 1.5 h at 37℃ in an orbital shaker at 75 rpm. After the initial adhesion phase, the cells suspensions were aspirated, and each well was washed twice with PBS to remove loosely adherent cells. Each well had 200 µL of fresh TSB added to promote biofilm growth and was incubated at 37℃ for 72 h. The amount of biofilm formed was measured using the XTT assay. Absorbance at 490 nm was measured following a 3 h incubation with XTT (1 mg/mL)-Menadion (0.4 mM). Presented values are mean±SD of three independent experiments. PBS, phosphate-buffered saline; TSB, tryptic soy broth.
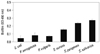
Fig. 2
The effect of co-culture of bacteria and C. albicans on biofilm formation. (A) Biofilm formation ability of C. albicans 53 was low. (B) Biofilm formation ability of C. albicans 163 was high. C. albicans (OD. 0.2) was cultured alone. Suspensions of bacteria (OD. 0.1) and C. albicans (OD. 0.1) were added to wells in a 96-well microtiter plate. The plate was incubated for 1.5 h at 37℃ in an orbital shaker at 75 rpm. After the initial adhesion phase, the cells suspensions were aspirated, and each well was washed twice with PBS to remove loosely adherent cells. The plate was incubated for 72 h at 37℃ in an orbital shaker at 75 rpm. The amount of biofilm formed was measured using the XTT assay. Absorbance at 490 nm was measured following incubation with XTT (1 mg/mL)Menadion (0.4 mM) for 3 h. Open bar: live C. albicans+live bacteria, black bar: C. albicans alone. Presented values are mean±SD of three independent experiments. p<0.05 was considered statistically significant. *p<0.05, **p<0.01. PBS, phosphate-buffered saline.

Fig. 3
The effect of the presence of dead bacteria on biofilm formation of C. albicans. (A) Biofilm formation ability of C. albicans 53 was low. (B) Biofilm formation ability of C. albicans 163 was high. The bacteria were killed by an incubation of 100℃ for 30 min. Suspensions of bacteria (OD. 0.1) and C. albicans (OD. 0.1) were added to wells in a 96-well microtiter plate. The plate was incubated for 1.5 h at 37℃ in an orbital shaker at 75 rpm. After the initial adhesion phase, the cells suspensions were aspirated, and each well was washed twice with PBS to remove loosely adherent cells. The plate was incubated for 72 h at 37℃ in an orbital shaker at 75 rpm. The amount of biofilm formed was measured using the XTT assay. Absorbance at 490 nm was measured following incubation with XTT (1 mg/mL)-Menadion (0.4 mM) for 3 h. Open bar: live C. albicans+dead bacteria, black bar: live C. albicans alone. Presented values are mean±SD of three independent experiments. p<0.05 was considered statistically significant. *p<0.05, **p<0.01. PBS, phosphate-buffered saline.
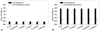
Fig. 4
The effect of the presence of dead C. albicans on the biofilm formation of bacteria. (A) Biofilm formation ability of C. albicans 53 was low. (B) Biofilm formation ability of C. albicans 163 was high. The C. albicans isolates were killed by incubation at 100℃ for 30 min. Suspensions of bacteria (OD. 0.1) and C. albicans (OD. 0.1) were added to wells in a 96-well microtiter plate. The plate was incubated for 1.5 h at 37℃ in an orbital shaker at 75 rpm. After the initial adhesion phase, the cells suspensions were aspirated, and each well was washed twice with PBS to remove loosely adherent cells. The plate was incubated for 72 h at 37℃ in an orbital shaker at 75 rpm. The amount of biofilm formed was measured using the XTT assay. The absorbance at 490 nm was measured following a 3 h incubation with XTT (1 mg/mL)-Menadion (0.4 mM). Open bar: dead C. albicans+live bacteria, black bar: live bacteria alone. Presented values are mean±SD of three independent experiments. PBS, phosphate-buffered saline.
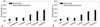
Fig. 5
Scanning electron micrograph images of monospecies (C. albicans or E. coli) and dual species (C. albicans and E. coli). (A) C. albicans 163 monospecies biofilm. (B) Live C. albicans 163 and live E. coli dual species biofilm. (C) Live C. albicans 163 and dead E. coli dual species biofilm. Magnifications are 500×(scale bar; 20 µm) and 4000×(scale bar; 2 µm), respectively.

Fig. 6
Relative quantitation of hyphae-specific genes (Als3, Ece1, Hwp1, Sap5) expression. The expressions of mRNA were evaluated via quantitative real-time reverse transcriptase polymerase chain reaction in C. albicans 163 biofilm formation. Data represent the mean±SD of three separate cultures. p<0.05 was considered statistically significant. *p<0.05, **p<0.01.

Fig. 7
Effect of live bacteria presence on expression of hyphae related gene on C. albicans. Relative quantitation of Als3 (A), Ece1 (B), Hwp1 (C), and Sap5 (D) gene expression was evaluated. C. albicans 163 cells were co-cultured with bacteria for 24 h and the target genes were determined by quantitative real-time reverse transcriptase polymerase chain reaction. Housekeeping gene Act1 and Pma1 were used for normalization. The data represent the average and standard deviation of three separate cultures. p<0.05 was considered statistically significant. *p<0.05, **p<0.01.

ACKNOWLEDGEMENTS
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2012R1A1A4A01011950).
References
1. Samaranayake LP. Essential microbiology for dentistry. 3rd ed. New York: Churchill Livingstone Elsevier;2006.
3. Ramage G, Martínez JP, López-Ribot JL. Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res. 2006; 6:979–986.

4. Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001; 183:5385–5394.

5. Nett J, Andes D. Candida albicans biofilm development, modeling a host-pathogen interaction. Curr Opin Microbiol. 2006; 9:340–345.

6. Baillie GS, Douglas LJ. Role of dimorphism in the development of Candida albicans biofilms. J Med Microbiol. 1999; 48:671–679.

7. Kojic EM, Darouiche RO. Candida infections of medical devices. Clin Microbiol Rev. 2004; 17:255–267.
8. Hansen SK, Rainey PB, Haagensen JA, Molin S. Evolution of species interactions in a biofilm community. Nature. 2007; 445:533–536.

9. Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005; 43:5721–5732.

10. Manson JM, Rauch M, Gilmore MS. The commensal microbiology of the gastrointestinal tract. Adv Exp Med Biol. 2008; 635:15–28.

11. Mclean RJC, Banes MB, McGowin CL, Aron GM. Methods of studying biofilms. In : Ghannoum MA, O'Toole GA, editors. Microbial biofilms. Washington, DC: ASM Press;2004. p. 379–413.
12. Tolker-Nielsen T, Hansen SK. Microbial interaction in mixed-species biofilms. In : Ghannoum MA, O'Toole GA, editors. Microbial biofilms. Washington, DC: ASM Press;2004. p. 206–221.
13. Shirtliff ME, Peters BM, Jabra-Rizk MA. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol Lett. 2009; 299:1–8.
14. Ramage G, Vandewalle K, Wickes BL, López-Ribot JL. Characteristics of biofilm formation by Candida albicans. Rev Iberoam Micol. 2001; 18:163–170.
15. Thein ZM, Seneviratne CJ, Samaranayake YH, Samaranayake LP. Community lifestyle of Candida in mixed biofilms: a mini review. Mycoses. 2009; 52:467–475.
16. El-Azizi MA, Starks SE, Khardori N. Interactions of Candida albicans with other Candida spp. and bacteria in the biofilms. J Appl Microbiol. 2004; 96:1067–1073.

17. Uppuluri P, Dinakaran H, Thomas DP, Chaturvedi AK, Lopez-Ribot JL. Characteristics of Candida albicans biofilms grown in a synthetic urine medium. J Clin Microbiol. 2009; 47:4078–4083.

18. Nailis H, Kucharíková S, Ricicová M, Van Dijck P, Deforce D, Nelis H, et al. Real-time PCR expression profiling of genes encoding potential virulence factors in Candida albicans biofilms: identification of model-dependent and -independent gene expression. BMC Microbiol. 2010; 10:114.

19. Costerton JW, Montanaro L, Arciola CR. Biofilm in implant infections: its production and regulation. Int J Artif Organs. 2005; 28:1062–1068.

20. Li X, Yan Z, Xu J. Quantitative variation of biofilms among strains in natural populations of Candida albicans. Microbiology. 2003; 149(Pt 2):353–362.

21. Hogan DA, Kolter R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science. 2002; 296:2229–2232.

22. Hogan DA, Vik A, Kolter R. A Pseudomonas aeruginosa quorum sensing molecule influences Candida albicans morphology. Mol Microbiol. 2004; 54:1212–1223.

23. Bandara HM, Yau JY, Watt RM, Jin LJ, Samaranayake LP. Escherichia coli and its lipopolysaccharide modulate in vitro Candida biofilm formation. J Med Microbiol. 2009; 58(Pt 12):1623–1631.

24. Staab JF, Bradway SD, Fidel PL, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 1999; 283:1535–1538.

26. Birse CE, Irwin MY, Fonzi WA, Sypherd PS. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect Immun. 1993; 61:3648–3655.

27. Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003; 67:400–428.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download