Abstract
Purpose
This study was conducted to investigate the small double-stranded RNA (dsRNA) mediated anti-tumor effects of p21WAF1/ClP1 (p21) transcriptional activation in vitro in the human glioma SHG-44 cell line.
Materials and Methods
Human glioma SHG-44 cells were transfected with dsRNA using LipofectAMINE 2000 transfection reagent. Real-time PCR and Western blot analysis were conducted to detect p21 and survivin mRNA and protein levels, respectively. Cell proliferation was examined by MTT assay. Cell cycle distribution and apoptosis were detected by flow-cytometric analysis.
Results
We found that dsRNA targeting p21 promoter (dsP21) significantly induced the expression of p21 at transcription and protein levels, and reduced the expression of survivin. AS well, dsP21 transcription significantly inhibited human glioma SHG-44 cell proliferation. Analysis of cell cycle distribution revealed that dsP21 transfection increased accumulation of cells in the G0/G1 phase and reduced accumulation of cells in the S phase. Further analysis revealed that dsP21 transcription led to an increase in both early and late stages of apoptosis in human glioma SHG-44 cells.
Glioma is the most common type of malignancy that originates in the central nervous system. Some of these tumors are highly malignant and tend to spread and infiltrate into normal nerve tissue, which makes surgical removal very difficult. Moreover, radiotherapy and chemotherapy are not sensitive to these tumors. The prognosis for patients with high-grade gliomas is generally poor, especially for older patients. Survival rates are 42.4% at 6 months, 17.7% at 1 year, and 3.3% at 2 years in these patients, according to a population-based study.1 Therefore, determining the pathogenesis of glioma and finding new methods are essential for improved clinical treatment of gliomas.
RNA-induced gene activation (RNAa) is a new mechanism of gene activation directed by small double-stranded RNA (dsRNA).2-5 dsRNA, are also referred to as 'small activating RNA' (saRNA) to distinguish them from small interfering RNA.6 By targeting gene promoter regions, saRNA induce the demethylation of histones, leading to transcriptional gene activation.7 Since the RNAa mechanism alters the chromatin structure leading to robust and prolonged expression of the endogenous target gene,2 it may be an attractive option to activate tumor suppressors in the treatment of cancer.
As a downstream mediator of tumor suppression, the p21 gene is linked to p53 expression and inhibition of cell cycle progression.8 It is involved in cell growth, differentiation, aging and death processes, and closely related to tumorigenesis. The p21 protein binds to cyclin-CDK2 or -CDK4 complexes and inhibits their activity. It is also an important regulatory protein of cell cycle progression. Previous studies have shown that decreased p21 expression may be involved in tumorigenesis or leads to poor prognosis of malignancy.9-11 Although prior experiments have shown the anti-tumor effects of p21 activation via RNAa in many human cell lines,12-15 on study has been done in human glioma cell lines.
Survivin is an inhibitor of the apoptosis protein family and has been implicated in anti-apoptosis, cell division, and cell cycle control.16,17 One previous study has reported that survivin and p21 are functionally associated with each other.18
Therefore, in this study, we attempted to investigate the anti-tumor effects of RNAa in human glioma SHG-44 cells and to examine survivin expression after dsP21 mediated p21 gene activation.
The design of dsRNA was performed as described previously by Li, et al.2 dsRNA targeting the p21 promoter at position 322 relative to the transcription start site [sense, 5'-CCAACUCAUUCUCCAAGUA(dT)(dT)-3'; antisense, 5'-UACUUGGAGAAUGAGUUGG(dT)(dT)-3'] was used to activate p21 expression. Control dsRNA (dsControl) lacking significant homology with any other human sequences (sense, 5'-UUCUCCGAACGUGUCACGUTT-3'; antisense, 5'-ACGUGACGUUCGGAGAATT-3) was used as a nonspecific control in this study. Synthetic dsRNA were green fluorescently-labeled and manufactured by Genepharma Company, Ltd. (Shanghai, China).
The human glioma cell line SHG-44 was purchased from the cell bank of China (Shanghai, China). SHG-44 cells were maintained in RPMI-1640 medium supplemented with penicillin G (100 U/mL), streptomycin (100 µg/mL), 2 mmol/L L-glutamine, and 10% fetal bovine serum. The cell line was incubated in a 37℃, 5% CO2 humidified incubator. The culture medium was changed every 48 h. The day before transfection, cells were plated in growth medium without antibiotics at a density of 50% to 60% (1×105/mL). Transfection of saRNA at a concentration of 50 nmol/L was carried out using LipofectAMINE 2000 reagent (Invitrogen, CA, USA) according to the manufacturer's instructions.
Total RNA was isolated from the cell by the standard Trizol method (RNAiso Plus, TaKaRa, Dalian, China). RNA (500 ng) was used for cDNA synthesis using the PrimeScript® RT Master Mix Perfect Real Time (TaKaRa). The resulting cDNA was amplified by PCR using gene-specific primers. p21 primers (sense, 5'-CATGTGGACCTGTCACT GTCTTGTA-3'; antisense, 5'-GAAGATCAGCCGGC GTTTG-3') and survivin primers (sense, 5'-GTCTGGCG TAAGATGATGGATTTG-3'; antisense, 5'-CACAGC AGTGTTTGAAATGACAGG-3') were used for real-time PCR analysis. PCR amplification included an initial denaturation step (95℃ for 30 s), 40 cycles of denaturation (95℃ for 5 s), annealing (60℃ for 30 s), and dissociation (95℃ for 15 s, 60℃ for 30 s, 95℃ for 15 s). Synthetic primers were manufactured by TaKaRa.
Cells were washed with ice-cold phosphate-buffered saline (PBS) at 72 h after transfection and lysed with RIPA Buffer (Pierce, MA, USA). Cell lysates were clarified by centrifugation at 12000×g for 30 min at 4℃ and protein concentrations were determined by using the BCA protein assay reagent (Pierce, MA, USA). Cell lysates were added to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, separated by SDS-PAGE and electrophoretically transferred to polyvinylidene difluoride membranes (Solarbio, Beijing, China). The membrane was detected with anti-p21 or antisurvivin antibodies (1:500; Bioworld Technology, Nanjing, China) and incubated at 4℃ overnight. Next, primary antibodies were removed and the membrane was detected by horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (1:10000; Bioworld Technology, Nanjing, China) and enhanced chemiluminescence detection (ECL System, Pierce, MA, USA).
Cells were transfected with dsRNA for approximately 6 h. Following treatments, cells were plated in 96-well microplates at a density of 3000 cells in 200 µL of complete RPMI-1640 medium per well for proliferation assay. Every 24 h, a batch of cells were stained with 20 µL of MTT [3-(4, 5-Dimethyl-2-thiazolyl)-2, 5-diphenyl-2Htetrazolium bromide] dye (5 mg/mL) at 37℃ for 4 h, after which the culture medium was removed and 100 µL of dimethyl sulfoxide was added and entirely mixed in for 10 minutes. Spectrometric absorbance at 490 nm was surveyed using a microplate reader.
Cells were plated in six-well plates at a density of 1×105 cells/mL. The next day, transfection was carried out and cells were incubated for 6 h before changing the transfection medium to fresh medium. Cells were harvested with trypsinization at 72 h, washed twice with pre-cooled PBS, fixed with cold 75% ethanol, and stained by propidium iodide (PI) in PBS. PI fluorescence intensity was surveyed by flow cytometry to assess cellular DNA content. Cells in the G0-G1, S, G2/M phases of the cell cycle were determined from the flow cytometry data. Apoptosis assays were also conducted to analyse the effect of p21 activation in SHG-44 cells with an Annexin V-fluorescein isothiocyanate apoptosis assay kit (Baiao Bioengineering Co. Ltd., Beijing, China). Transfected cells were harvested, washed with pre-cooled PBS twice, resuspended in binding buffer, and stained by Annexin V and PI according to the manufacturers' instructions. Annexin V stained cells indicate early apoptotic cells, whereas Annexin V+PI stained cells indicate late apoptotic cells. All of the samples were assayed in triplicate.
Cells were plated in six-well plates at a density of 1×105 cells/mL and washed with ice-cold PBS twice at 72 h after transient transfection. Then, total cell numbers and fluorescent cells in the same field were counted respectively by phase contrast microscope, and transfection efficiency was calculated according to the following formula: fluorescent cells/total cell numbers×100%. The transfection efficiency was 57.2% (Fig. 1).
Previous studies have demonstrated that dsRNA targeting the p21 gene promoter at position 322, relative to the transcription start site, can activate p21 expression.2,14 In the present study, SHG-44 cells were transiently transfected with 50 nmol/L of dsP21 and a nonspecific control dsRNA for 72 h, and expression of p21 mRNA and protein was evaluated by real-time PCR and Western blotting, respectively. Expression of p21 mRNA in dsP21-transfected cells was significantly elevated compared to mock and dsControl treatments (Fig. 2A). Induction of p21 was also confirmed by Western blot analysis (Fig. 2B and C). As well, elevated surlevels of p21 protein were strongly correlated with increases in p21 mRNA expression in SHG-44 cells.
Because up-regulation of p21 leads to an inhibition of tumor growth, we thus examined the effect of p21 transcriptional activation on the proliferation of glioma SHG-44 cells in vitro. In this experiment, cellular proliferation was monitored by MTT assay daily for 6 days. Cell growth curve showed that, compared with mock and dsControl treatments, dsP21 transfected cells were significantly inhibited in a time-dependent manner, while dsControl and mock cells showed no significant inhibition of proliferation (Fig. 3).
Apoptosis assays were used to investigate the effect of p21 knockdown on the growth of human glioma SHG-44 cells. The early and the late apoptosis rate of dsP21-transfected cells significantly increased compared to the mock and dsControl treatments, while there were no differences in apoptosis rates among the latter two types of cells (Fig. 4). Increases in both early and late apoptosis rates were seen.
Cell cycle analysis was conducted to investigate the cell cycle distribution of dsP21-transfected glioma SHG-44 cells. The percentage of the cells in the G0/G1 phase was statistically significant increased in the dsP21-transfected cells compared to those in the mock and dsControl cells. Transfection with dsP21 also caused a decrease in S-phase cells, but no change was found in G2/M phase cell populations (Fig. 5).
Real-time PCR and western blot analysis were conducted to examine the effect of p21 activation on survivin mRNA and protein. As shown in Fig. 6A, compared to mock and dsControl cells, a statistically significant decrease in survivin mRNA was observed when transfected with dsP21 (Fig. 6A). The decrease in survivin protein was further evaluated by Western blot analysis. The expression of survivin protein was significantly decreased in dsP21 transfected cells compared with both mock and dsControl treatments (Fig. 6B and C).
A tumor suppressor gene is a gene that protects a cell from one step on the path to cancer. Inactivation of tumor suppressor genes is an important cause of tumorigenesis. Gene mutation, deletion, and structural chromosomal rearrangements are an important mechanism for the inactivation of tumor suppressor genes.19
Previous studies have already confirmed that inactivation of p21 expression may be involved in tumorigenesis or lead to poor prognosis of malignancy.9-11 Interestingly, other findings have found that increased p21 expression is associated with tumor progression or worse prognosis.20-23 These studies suggest that p21 may act as an oncogene, either during tumor development or in the course of anti-cancer treatment.
Accordingly, there are questions as to whether p21 is a Retumor suppressor or an oncogene. This discrepancy could be due to the status of p21 itself and/or to differences in the histological types of cancers that have been analyzed.24 Hukkelhoven, et al.25 confirmed that tyrosine phosphorylation contributes to the conversion of cdk inhibitors from tumor suppressive roles to oncogenic roles. Besson, et al.26 reported that control of the subcellular localization of p21 could represent an important regulatory switch from a nuclear tumor suppressor to a cytoplasmic oncogene. In the current study, we demonstrated that p21 plays a tumor suppressive role in human glioma cell lines and that it may be a potentially desirable target for glioma treatment.
Many studies have reported that use of dsRNA targeting gene promoters to activate expression of tumor suppressor genes thereby inhibits tumor cell proliferation and migration, leading to cell cycle arrest and induction of apoptosis.2,7,13,27 Matsui, et al.28 reported that implementing duplex RNA complementary to the promoter of LDL receptor (LDLR) activated expression of LDLR and increased the display of LDLR on the surface of liver cells. Additionally, Chen, et al.29 utilized ribonucleic acid RNAa mechanisms to increase the expression of VEGF to treat erectile function. As recent studies have suggested that RNAa depends on Argonaute (AGO) proteins, Chu, et al.30 investigated the role of AGO1-4 in gene silencing and activation of the progesterone receptor gene. Their data indicated that expression of AGO2 is necessary for efficient gene silencing or activation: saRNA loading and processing by an AGO protein, which then guides it to its promoter target, which can be a non-coding transcript overlapping the promoter or the chromosomal DNA, and recruits histone modifying enzymes to the promoter to activate transcription by causing permissive epigenetic changes.6,28,30
Small saRNA mediated gene activation offers a promising new approach for investigating gene function, and may serve as a novel strategy for the treatment of many diseases, especially for tumors. We designed this experiment to examine whether induction of p21 by RNAa has an anti-tumor effect on human glioma cells as an effort to explore novel therapeutic strategies for the treatment of human gliomas. In our study, we found the transfection efficiency of SHG-44 cells to be satisfactory, and activation of gene expression by RNAa may be a likely therapy strategy for the treatment of gliomas. After transfection with dsP21 into SHG-44 cells at 72 h, the expression of p21 in SHG-44 cells was significantly increased, compared to mock and dsControl treatments, according to real time PCR and Western blotting results. Furthermore, induction of p21 protein expression led to a significant inhibition of SHG-44 cells proliferation. Moreover, expression of p21 upregulation induced the accumulation of cells in the G0/G1 phase and significantly increased the early and late apoptosis rates of dsP21-transfected cells.
RNAa-mediated overexpression of p21 in human glioma SHG-44 cells suppressed expression of survivin. Hence, this result suggests that survivin may serve as a downstream factor of p21 to promote cell cycle arrest and enhance apoptosis.
In conclusion, the preset study demonstrated dsRNA-mediated gene activation in a human glioma cell line. Induction of p21 by RNAa exhibited anti-tumor activity in vitro in glioma SHG-44 cells by inhibitting cell cycle progression and inducing apoptosis. Further research should focus on revealing the exact mechanism of RNAa and develop potent reagents for laboratory and clinical therapeutic application.
Figures and Tables
Fig. 1
The transfection efficiency of human glioma SHG-44 cells. The transfection efficiency was 57.2% (magnification×10). (A) Inverted phase contrast microscope Bright field 10×. (B) Inverted phase contrast microscope Darkfield 10×.

Fig. 2
dsP21 induces p21 expression in glioma SHG-44 cells. Induction of p21 mRNA expression was detected by real-time PCR, p21 mRNA expression levels were normalized to mock and calculated by 2-ΔΔCt, and the results were presented as means±SD of three independent experiments. Compared to mock, there was a two-fold induction in dsP21 mRNA expression (mock vs. dsControl: p=0.400, p>0.05; mock vs. dsP21, p=0.001, p<0.05; dsControl vs. dsP21, p=0.000, p<0.05) (A). In addition, p21 protein expression levels were detected by Western blot analysis. β-actin levels were detected as an endogenous control. p21 protein expression levels were normalized to β-actin and presented as the means±SD of three independent experiments (p<0.05, mock: 0.426±0.010, dsControl: 0.400±0.005 vs. dsP21: 0.954±0.030) (B and C). *Compared to Mock and dsControl, p<0.05.

Fig. 3
Upregulation of p21 leads to an inhibition of SHG-44 cell proliferation in a time-dependent manner. The spectrometric absorbance at 490 nm of dsP21 transfected glioma cells significantly decreased to 0.134±0.035, 0.138±0.038, 0.150±0.05, 0.178±0.03 at 3, 4, 5, and 6 days, compared to mock and dsControl (p<0.05, mock: 0.225±0.063, 0.367±0.057, 0.375±0.049, 0.609±0.051; dsControl: 0.233±0.061, 0.336±0.044, 0.393±0.066, 0.627±0.066). (A) Cell growth curve. (B) Histogram. *Compared to Mock and dsControl, p<0.05.

Fig. 4
dsP21 treatment induced apoptosis of human glioma SHG-44 cells. The early and the late apoptosis rates of dsP21 transfected cells increased to 8.14% and 50.0%, compared to the mock and dsControl treatments (early apoptosis rate, p<0.05, 8.14% vs. 2.11%, 2.37%), (late apoptosis rate, p<0.05, 50.0% vs. 11.7%, 13.9%), respectively. Meanwhile, there were no obvious differences in apoptosis rates between the latter two types of cells (the early apoptosis rate, p>0.05, 2.11% vs. 2.37%; the late apoptosis rate, p>0.05, 11.7% vs. 11.9%). (A) Apoptosis detection. (B) Histogram. *Compared to Mock and dsControl, p<0.05. 1, necrosis cells; 2, late apoptotic cells; 3, live cells; 4, early apoptotic cells.
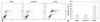
Fig. 5
Effect of dsP21 treatment on cell cycle distributions of glioma SHG-44 cells. The percentage of cells in the G0/G1 phase significantly increased to 72.29% in the dsP21-transfected cells, compared to those in the mock and dsControl cells (p<0.05, 72.29% vs. 67.52%, 64.88%), while there were no obvious differences between the latter two types of cells (p>0.05, 67.52%, 64.88%). Furthermore, a decrease in the S-phase fraction was seen in the dsP21-transfected cells, compared to those in the mock and dsControl cells (p<0.05, 10.26% vs. 14.83%, 19.67%). No change was seen in the G2/M phase cell populations (p>0.05, 17.45% vs. 17.65%, 15.45%). (A) Cell cycle analysis. (B) Histogram. *Compared to Mock and dsControl, p<0.05.

Fig. 6
dsP21 transfection is associated with a decrease in survivin expression in human glioma SHG-44 cells. Induction of survivin mRNA expression was detected by real-time PCR; survivin mRNA expression levels were normalized to mock and calculated by 2-ΔΔCt, and the results were presented as means±SD of three independent experiments. Compared to mock, a two-fold reduction in survivin mRNA expression occurred in dsP21 transfected cells (mock vs. dsControl: p=0.562, p>0.05; mock vs. dsP21, p=0.000, p<0.05; dsControl vs. dsP21, p=0.007, p<0.05) (A). In addition, survivin protein expression levels were detected by Western blot analysis. β-actin levels were detected as an endogenous control. Survivin protein expression levels were normalized to β-actin and presented as the means±SD of three independent experiments (p<0.05, mock: 0.562±0.051, dsControl: 0.543±0.012 vs. dsP21: 0.193±0.009) (B and C).

ACKNOWLEDGEMENTS
We would like to thank Yinian Zhang, MD, who helped revise the English writing. The work was performed at the Medical Center Laboratory of Lanzhou University.
References
1. Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004; 64:6892–6899.
2. Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A. 2006; 103:17337–17342.

3. Huang V, Qin Y, Wang J, Wang X, Place RF, Lin G, et al. RNAa is conserved in mammalian cells. PLoS One. 2010; 5:e8848.

4. Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008; 105:1608–1613.

5. Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004; 116:779–793.

6. Portnoy V, Huang V, Place RF, Li LC. Small RNA and transcriptional upregulation. Wiley Interdiscip Rev RNA. 2011; 2:748–760.

7. Junxia W, Ping G, Yuan H, Lijun Z, Jihong R, Fang L, et al. Double strand RNA-guided endogeneous E-cadherin up-regulation induces the apoptosis and inhibits proliferation of breast carcinoma cells in vitro and in vivo. Cancer Sci. 2010; 101:1790–1796.

8. Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995; 55:5187–5190.
9. Shi YZ, Hui AM, Takayama T, Li X, Cui X, Makuuchi M. Reduced p21(WAF1/CIP1) protein expression is predominantly related to altered p53 in hepatocellular carcinomas. Br J Cancer. 2000; 83:50–55.

10. Goto M, Tsukamoto T, Inada K, Mizoshita T, Ogawa T, Terada A, et al. Loss of p21WAF1/CIP1 expression in invasive fronts of oral tongue squamous cell carcinomas is correlated with tumor progression and poor prognosis. Oncol Rep. 2005; 14:837–846.

11. Shoji T, Tanaka F, Takata T, Yanagihara K, Otake Y, Hanaoka N, et al. Clinical significance of p21 expression in non-small-cell lung cancer. J Clin Oncol. 2002; 20:3865–3871.

12. Wei J, Zhao J, Long M, Han Y, Wang X, Lin F, et al. p21WAF1/CIP1 gene transcriptional activation exerts cell growth inhibition and enhances chemosensitivity to cisplatin in lung carcinoma cell. BMC Cancer. 2010; 10:632.

13. Whitson JM, Noonan EJ, Pookot D, Place RF, Dahiya R. Double stranded-RNA-mediated activation of P21 gene induced apoptosis and cell cycle arrest in renal cell carcinoma. Int J Cancer. 2009; 125:446–452.

14. Chen Z, Place RF, Jia ZJ, Pookot D, Dahiya R, Li LC. Antitumor effect of dsRNA-induced p21(WAF1/CIP1) gene activation in human bladder cancer cells. Mol Cancer Ther. 2008; 7:698–703.

15. Wu ZM, Dai C, Huang Y, Zheng CF, Dong QZ, Wang G, et al. Anti-cancer effects of p21WAF1/CIP1 transcriptional activation induced by dsRNAs in human hepatocellular carcinoma cell lines. Acta Pharmacol Sin. 2011; 32:939–946.

16. Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997; 3:917–921.

17. Suzuki A, Shiraki K. Tumor cell "dead or alive": caspase and survivin regulate cell death, cell cycle and cell survival. Histol Histopathol. 2001; 16:583–593.
18. Suzuki A, Ito T, Kawano H, Hayashida M, Hayasaki Y, Tsutomi Y, et al. Survivin initiates procaspase 3/p21 complex formation as a result of interaction with Cdk4 to resist Fas-mediated cell death. Oncogene. 2000; 19:1346–1353.

19. Zhang Z, Li D, Wu M, Xiang B, Wang L, Zhou M, et al. Promoter hypermethylation-mediated inactivation of LRRC4 in gliomas. BMC Mol Biol. 2008; 9:99.

20. Ferrandina G, Stoler A, Fagotti A, Fanfani F, Sacco R, De Pasqua A, et al. p21WAF1/CIP1 protein expression in primary ovarian cancer. Int J Oncol. 2000; 17:1231–1235.

21. Cheung TH, Lo KW, Yu MM, Yim SF, Poon CS, Chung TK, et al. Aberrant expression of p21(WAF1/CIP1) and p27(KIP1) in cervical carcinoma. Cancer Lett. 2001; 172:93–98.

22. Rau B, Sturm I, Lage H, Berger S, Schneider U, Hauptmann S, et al. Dynamic expression profile of p21WAF1/CIP1 and Ki-67 predicts survival in rectal carcinoma treated with preoperative radiochemotherapy. J Clin Oncol. 2003; 21:3391–3401.

23. Sarbia M, Gabbert HE. Modern pathology: prognostic parameters in squamous cell carcinoma of the esophagus. Recent Results Cancer Res. 2000; 155:15–27.

24. Xia X, Ma Q, Li X, Ji T, Chen P, Xu H, et al. Cytoplasmic p21 is a potential predictor for cisplatin sensitivity in ovarian cancer. BMC Cancer. 2011; 11:399.

25. Hukkelhoven E, Liu Y, Yeh N, Ciznadija D, Blain SW, Koff A. Tyrosine phosphorylation of the p21 cyclin-dependent kinase inhibitor facilitates the development of proneural glioma. J Biol Chem. 2012; 287:38523–38530.

26. Besson A, Assoian RK, Roberts JM. Regulation of the cytoskeleton: an oncogenic function for CDK inhibitors. Nat Rev Cancer. 2004; 4:948–955.

27. Wang J, Place RF, Huang V, Wang X, Noonan EJ, Magyar CE, et al. Prognostic value and function of KLF4 in prostate cancer: RNAa and vector-mediated overexpression identify KLF4 as an inhibitor of tumor cell growth and migration. Cancer Res. 2010; 70:10182–10191.

28. Matsui M, Sakurai F, Elbashir S, Foster DJ, Manoharan M, Corey DR. Activation of LDL receptor expression by small RNAs complementary to a noncoding transcript that overlaps the LDLR promoter. Chem Biol. 2010; 17:1344–1355.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download