Abstract
Purpose
Materials and Methods
Results
Figures and Tables
Table 2

SBT, sequence-based typing; HLA, human leukocyte antigen; IMGT, international immunogenetics.
*HLA genotypes were determined based on the IMGT/HLA database release 3.9.0. The occurrence of ambiguous genotyping results and the number of ambiguous allele combinations by an SBT assay for an individual may change according to the version of the IMGT/HLA database release.
†Average number of loci with ambiguity per sample was 3.28 in the 50 specimens, and this was reduced to 1.22 (61.8% reduction) with the AVITA plus assay. A total of 48 (96.0%) cases among the 50 specimens showed reduction in the number of ambiguous allele combinations for any of the four HLA loci.
Table 3

SBT, sequence-based typing; HLA, human leukocyte antigen; IMGT, international immunogenetics.
*HLA genotypes were determined based on the IMGT/HLA database release 3.9.0. The occurrence of ambiguous genotyping results and the number of ambiguous allele combinations by an SBT assay for an individual may change according to the version of the IMGT/HLA database release.
†Expected genotype frequency was calculated by the Hardy-Weinberg equation with previously reported allele frequencies in Korean and US populations.18,20
Table 4

SBT, sequence-based typing; HLA, human leukocyte antigen; IMGT, international immunogenetics.
*HLA genotypes were determined based on the IMGT/HLA database release 3.9.0. The occurrence of ambiguous genotyping results and the number of ambiguous allele combinations by an SBT assay for an individual may change according to the version of the IMGT/HLA database release.
†Expected genotype frequency was calculated by the Hardy-Weinberg equation with previously reported allele frequencies in Korean and US populations.18,20
Table 5
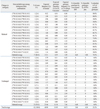
SBT, sequence-based typing; HLA, human leukocyte antigen; IMGT, international immunogenetics.
*HLA genotypes were determined based on the IMGT/HLA database release 3.9.0. The occurrence of ambiguous genotyping results and the number of ambiguous allele combinations by an SBT assay for an individual may change according to the version of the IMGT/HLA database release.
†Expected genotype frequency was calculated by the Hardy-Weinberg equation with previously reported allele frequencies in Korean and US populations.18,20
Table 6

SBT, sequence-based typing; HLA, human leukocyte antigen; IMGT, international immunogenetics.
*HLA genotypes were determined based on the IMGT/HLA database release 3.9.0. The occurrence of ambiguous genotyping results and the number of ambiguous allele combinations by an SBT assay for an individual may change according to the version of the IMGT/HLA database release.
†Expected genotype frequency was calculated by the Hardy-Weinberg equation with previously reported allele frequencies in Korean and US populations.18,20




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download