Abstract
Purpose
The increasing prevalence and global spread of carbapenem-resistant Acinetobacter baumannii (A. baumannii) has become a serious problem. The aim of this study was to investigate molecular and epidemiological characteristics of carbapenem-resistant A. baumannii isolates collected from Korean non-tertiary hospitals.
Materials and Methods
Thirty six non-duplicated carbapenem-resistant A. baumannii isolates were collected from 17 non-tertiary hospitals in Korea between 2004 and 2006. Isolates were typed by multilocus sequence typing and repetitive-sequence-based PCR (rep-PCR). Detection of genes encoding OXA carbapenemase and their relationship with ISAba1 was performed by PCR.
Results
Two clones were prevalent among 36 isolates: ST69 (17 isolates, 47.2%) and ST92 (19 isolates, 52.8%). Rep-PCR patterns were diverse and revealed that all isolates were clustered into eight band patterns. The ISAba1-activated blaOXA-23-like and ISAba1-activated blaOXA-51-like genes were prevalent among the carbapenem-resistant A. baumannii isolates.
Acinetobacter baumannii (A. baumannii) has emerged worldwide as an important opportunistic nosocomial pathogen, especially in intensive care unit (ICU) patients.1 Carbapenems have been widely used to treat serious infections associated with multidrug-resistant (MDR) A. baumannii, but carbapenem resistance has been increasingly reported worldwide.2,3 Moreover, most carbapenem-resistant A. baumannii strains are multidrug or even pandrug resistant.4 In Korea, imipenem-resistant A. baumannii isolates from a university hospital reached 32.5%.5 Among the various mechanisms of carbapenem resistance in A. baumannii, carbapenemase of the molecular class D OXA enzymes has emerged as the main source of carbapenem resistance.6 A previous study reported that among ten university hospitals in Korea, 78.3% of carbapenem-resistant A. baumannii isolates were caused by OXA-23-like enzymes, among which ST22 was the most prevalent type.7 Park, et al.8 reported that two major clones of carbapenem-resistant A. baumannii, ST22 and ST28, were found in five tertiary care hospitals.8 However, these two clones have so far been studied only in tertiary hospital isolates. This study was performed to analyze the genetic diversity and clonal distribution of carbapenem-resistant A. baumannii isolates among non-tertiary hospitals in Korea.
A total of 315 consecutive A. baumannii clinical isolates were collected from 24 non-tertiary South Korea hospitals between 2004 and 2006. Among these isolates, 50 were carbapenem-resistant, of which 36 nonrepetitive isolates from 17 hospitals in 8 provinces were selected for this study. The strains were propagated at 37℃ in Luria-Bertani broth or agar. The clinical isolates were identified by using the Vitek II system (bioMerieux, Marcy-I'Etoile, France). Species identification was performed by partial rpoB gene sequencing.9
Antimicrobial susceptibility tests were performed using the minimal inhibitory concentrations of Etest (AB BIODISK, Piscataway, NJ, USA), based on the results reported by the Clinical and Laboratory Standards Institute.10 Eleven antimicrobial agents were tested: amikacin, gentamicin, ticarcillin-clavulanic acid, piperacillin, imipenem, meropenem, ciprofloxacin, ceftazidime, cefepime and polymyxin B, according to the manufacturer's instructions. Escherichia coli ATCC 25922 was used for quality control in antimicrobial susceptibility testing.
MLST was performed for all isolates according to the method described previously.11 Fragments of seven housekeeping genes (gltA, gyrB, gdhB, recA, cpn60, gpi, rpoD) were amplified by PCR and sequenced. Allele numbers were assigned as sequence types (STs) after distinct allele sequences were submitted to a dedicated data base (http://pubmlst.org). A clonal complex was used to assess the genetic relatedness of the STs; the most stringent definition-sharing alleles at 6 of 7 loci-was used.12
Rep-PCR was performed by using the DiversiLab system (bioMerieux SA, Grenoble, France) to investigate the clonal relatedness of A. baumannii isolates. Results were analyzed with DiversiLab software using the Kullback-Leibler method to determine distance matrices, and the unweighted pair group method with arithmetic averages to create a dendrogram. Sample relationships were designated as follows: pattern, no band differences (indistinguishable); group, 1 to 2 band differences (similar); and different group, 3 or more band differences. Each group corresponded to an outbreak.
PCR was performed to detect the genes encoding MBLs (IMP-1 variants and VIM-2 variants) and OXA carbapenemase (OXA-23-like, OXA-24-like, OXA-51-like and OXA-58-like) according to the method described previously.13 The sequence analysis of PCR products was carried out by request at Macrogen (http://macrogen.co.kr).
The isolated carbapenem-resistant A. baumannii isolates showed 100% resistance to gentamicin, piperacillin, ciprofloxacin, ceftazidime and cefepime. In addition, they showed a high resistance against amikacin (34/36, 94.4%), but a complete susceptibility to polymyxin B (Table 1).
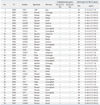
Two unique STs among the 36 carbapenem-resistant A. baumannii isolates were revealed by MLST analysis (Table 2). Two clones were prevalent among 36 isolates: ST69 (17 isolates, 47.2%) and ST92 (19 isolates, 52.8%). All 36 A. baumannii isolates carried the blaOXA-51-like gene. Fourteen isolates (38.9%) had the ISAba1-activated blaOXA-23-like gene, and 25 isolates (69.4%) had the ISAba1-activated blaOXA-51-like gene. Coexistence of blaOXA-23 and ISAba1-activated blaOXA-51-like genes was detected in three isolates (8.3%). No blaOXA-24-like, blaOXA-58 like, IMP-1 or VIM-2 genes were found.
The results of rep-PCR showed that carbapenem-resistant A. baumannii isolates were clustered into eight distinctive band patterns, including five clonally related groups. A 96.5% similarity line was obtained by DiversiLab software to represent the best match between pattern assignments and the dendrogram (Fig. 1). The following differences in rep-PCR patterns were observed between ST69 and ST92: band pattern 1 [2 isolates (ST69)], band pattern 2 [2 isolates (ST69)], band pattern 3 [5 isolates (ST69), 1 isolate (ST92)], band pattern 4 [3 isolates (ST69), 1 isolate (ST92)], band pattern 5 [1 isolate (ST69), 13 isolates (ST92)], band pattern 6 [4 isolates (ST92)], band pattern 7 [3 isolates (ST69)], and band pattern 8 [1 isolate (ST69)] (Table 2).
A. baumannii is a rapidly emerging pathogen in hospitals, frequently causing nosocomial outbreaks in ICUs. Several hospital outbreaks caused by MDR A. baumannii clones have been described in Europe and worldwide. The incidence of carbapenem-resistant A. baumannii has increased worldwide since its first emergence in New York in 1991.14 Despite the high ratio of carbapenem-resistant A. baumannii isolates in Korean tertiary hospitals,15 there have been no reports about the antimicrobial susceptibility, resistance mechanisms or molecular epidemiology of carbapenem-resistant A. baumannii in Korean non-tertiary hospitals. In this study, we used MLST and rep-PCR to investigate the clonality of carbapenem-resistant A. baumannii isolates in non-tertiary hospitals throughout various regions of Korea. In MLST, two distinctive clones, ST92 and its double-locus variant ST69, were identified among 36 carbapenem-resistant A. baumannii isolates. Our results corresponded well with those of an earlier study which reported that ST92 (ST22 was redesignated ST92 after an update of allelic profiles at the database website http://pubmist.org) may contribute to the high carbapenem resistance rates of Acinetobacter isolates in Korean university hospitals.7 Wide dissemination of non-carbapenem-susceptible ST92 isolates has also been described in China and Korea.16 In addition, Park, et al.8 demonstrated that ST69 was another major carbapenem-resistant clone found in tertiary care hospitals in Korea. ST28, however, was not identified in this study, which was previously indicated as a major carbapenem-resistant clone found in Korea.8 There were no distinctive characteristics in regards to blaOXA type in the two major clones, ST92 and ST69, as shown in Table 2. Because both clones were found in most regions, they may be distributed ubiquitously throughout Korea. Most of the isolates of the ST92 clone (17/19, 89.5%) were clustered into three rep-PCR band groups. The ISAba1-activated OXA-23-like β-lactamase gene was found in the isolates of ST69 (9 isolates) and ST92 (5 isolates).
In summary, we investigated the antimicrobial resistance and clonality of carbapenem-resistant A. baumannii isolates in non-tertiary hospitals throughout Korea. Carbapenem-resistant A. baumannii isolates in Korea may be due to two major clones, ST69 and ST92, which have disseminated nationwide in not only tertiary hospitals but also in non-tertiary hospitals throughout Korea.
Figures and Tables
ACKNOWLEDGEMENTS
This study was supported by an intramural research fund of the Korea Centers for Disease Control and Prevention. 2007-N44002-00.
References
2. Poirel L, Lebessi E, Héritier C, Patsoura A, Foustoukou M, Nordmann P. Nosocomial spread of OXA-58-positive carbapenem-resistant Acinetobacter baumannii isolates in a paediatric hospital in Greece. Clin Microbiol Infect. 2006. 12:1138–1141.

3. Garnacho-Montero J, Amaya-Villar R. Multiresistant Acinetobacter baumannii infections: epidemiology and management. Curr Opin Infect Dis. 2010. 23:332–339.

4. Kwon KT, Oh WS, Song JH, Chang HH, Jung SI, Kim SW, et al. Impact of imipenem resistance on mortality in patients with Acinetobacter bacteraemia. J Antimicrob Chemother. 2007. 59:525–530.

5. Lee JH, Choi CH, Kang HY, Lee JY, Kim J, Lee YC, et al. Differences in phenotypic and genotypic traits against antimicrobial agents between Acinetobacter baumannii and Acinetobacter genomic species 13TU. J Antimicrob Chemother. 2007. 59:633–639.

7. Park YK, Lee GH, Baek JY, Chung DR, Peck KR, Song JH, et al. A single clone of Acinetobacter baumannii, ST22, is responsible for high antimicrobial resistance rates of Acinetobacter spp. isolates that cause bacteremia and urinary tract infections in Korea. Microb Drug Resist. 2010. 16:143–149.

8. Park YK, Choi JY, Jung SI, Park KH, Lee H, Jung DS, et al. Two distinct clones of carbapenem-resistant Acinetobacter baumannii isolates from Korean hospitals. Diagn Microbiol Infect Dis. 2009. 64:389–395.

9. La Scola B, Gundi VA, Khamis A, Raoult D. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J Clin Microbiol. 2006. 44:827–832.

10. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twentieth Informational Supplement M100-S20. 2010. Wayne, PA, USA: CLSI.
11. Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodríguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005. 43:4382–4390.

12. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004. 186:1518–1530.

13. Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006. 27:351–353.

14. Manikal VM, Landman D, Saurina G, Oydna E, Lal H, Quale J. Endemic carbapenem-resistant Acinetobacter species in Brooklyn, New York: citywide prevalence, interinstitutional spread, and relation to antibiotic usage. Clin Infect Dis. 2000. 31:101–106.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download