Abstract
Autophagy is a specialized cellular pathway involved in maintaining homeostasis by degrading long-lived cellular proteins and organelles. Recent studies have demonstrated that autophagy is utilized by immune systems to protect host cells from invading pathogens and regulate uncontrolled immune responses. During pathogen recognition, induction of autophagy by pattern recognition receptors leads to the promotion or inhibition of consequent signaling pathways. Furthermore, autophagy plays a role in the delivery of pathogen signatures in order to promote the recognition thereof by pattern recognition receptors. In addition to innate recognition, autophagy has been shown to facilitate MHC class II presentation of intracellular antigens to activate CD4 T cells. In this review, we describe the roles of autophagy in innate recognition of pathogens and adaptive immunity, such as antigen presentation, as well as the clinical relevance of autophagy in the treatment of human diseases.
Autophagy is a part of cellular system involved in maintaining homeostasis by degrading long-lived cellular constituents.1 It also plays critical roles in providing nutrients under starvation and neonatal periods.2,3 There are three types of autophagy: macroautophagy, microautophagy, and chaperone-mediated autophagy.4 In chaperone-mediated autophagy, signaling motif containing molecules are transported with the chaperone HSC70 via LAMP-2A into lysosomes.5-8 In contrast to microautophagy, which is characterized by the removal of constituents via budding of an autophagic body at the lysosomal membrane, macroautophagy forms a double-layered membrane vesicle, called an autophagosome. The autophagosome is formed via the elongation of a cup-shaped membrane, and two ubiquitin-like conjugation systems are involved in autophagosome propagation.9 At least 30 genes, termed autophagy-related genes (Atg), regulate the process of autophagy in yeast.10 Once formed, the outer membrane of the autophagosome fuses with a lysosome, where cellular contents are degraded within by lysosomal hydrolase and recycled.11
Beyond maintaining homeostasis, autophagy is involved in multiple biological processes including development, aging, and degeneration.12 Not surprisingly, aberrant regulation of autophagy induces many diseases such as cancer, neurodegenerative disease, and myopathies.13,14 Autophagy also has diverse functions in immunity. Various intracellular bacteria, viruses, and protozoans are removed from host cells by autophagy, and endogenous antigens are processed and presented to major histocompatibility complex (MHC) class II via autophagy.15-21 In this review, we focus on the role of autophagy in innate recognition of pathogens and adaptive immune responses.
In response to pathogens, various types of pattern recognition receptors (PRRs) recognize pathogen-associated molecular patterns (PAMPs) and mediate signals to defend to pathogens.22 Among PRRs, Toll-like receptors (TLRs) respond to lipopolysaccharides (LPS), lipotechoic acid and flagellin on cell surface membranes, as well as to viral/bacterial nucleic acids on endosomal membranes.23 TLR4, a receptor for bacterial LPS, triggers both MyD88- and TIR domain-containing adapter-inducing interferon-β (TRIF)-dependent signaling pathways. The IKK-α-IKK-β-NEMO complex and TBK1-IKKi complex mediate the activation of the transcription factors NF-κB and interferon regulatory factor 3 (IRF3), respectively. In turn, they induce the transcription of proinflammatory cytokines and type I interferons (IFNs).24,25 TLR4 signaling via the TRIF-p38 axis, but not via MyD88, induces the formation of an autophagosome for the elimination of Mycobacteria bacilli.26 Atg6 and Beclin-1 are required in this process (Fig. 1A). Interestingly, in autophagy-deficient cells, IL-1β and IL-18 production is enhanced in response to LPS.27 Macrophages lacking Atg16L1 induce high-levels of reactive oxygen species (ROS), which in turn activates caspase-1, leading to the processing of IL-1β. However, in macrophages of wild-type mice, the generation of ROS is inhibited by autophagy-related proteins, and in turn, limited amounts of IL-1β are produced (Fig. 1C).
In addition to TLR4 signaling, other TLRs also activate autophagy machinery to eliminate pathogens. TLR7 signaling, induced by two different ligands, single-stranded RNA and imiquimod, induces the formation of autophagosomes, characterized by microtubule-associated light chain 3-green fluorescent protein (LC3-GFP) puncta formation for the elimination of Bacillus Calmette-Guerin.28,29 The induction of autophagy is dependent on MyD88. Here, both Atg5 and Beclin are required for the induction of autophagy in macrophages after stimulation of TLR7 (Fig. 1A).
Although most TLR signaling induces autophagosome formation, some link autophagy with the enhancement of phagocytosis. When zymosan (a particle of fungal cell walls) is phagocytosed, LC3, an autophagosome marker, is rapidly recruited to the phagosomal membrane.30 This process depends on Atg5 and Atg7. Although translocation of LC3 to the phagosome is not associated with autophagosome formation, it promotes the phagosome to fuse with lysosomes in murine macrophages. Interestingly, recruitment of LC3 to phagosome depends on TLR2, but not on MyD88 (Fig. 1A).
In addition to the formation of autophagosomes and maturation of phagosomes by autophagy-related proteins to promote TLR-mediated pathogen elimination, autophagy supports delivery of viral nucleic acids to lysosomes where TLR recognition occurs.31 Within the TLR family, TLR3, TLR7, TLR8 and TLR9, located in the endosomal compartment, sense endocytosed viral nucleic acids.32 In the case of vesicular stomatitis virus (VSV), plasmacytoid dendritic cells (pDCs) recognize replication intermediates in the cytosol, rather than single-stranded viral RNA via TLR7 in the endosome.31 In recognition of cytosolic viral PAMP by TLR7, autophagy mediates the transport thereof into lysosomes (Fig. 1B). Consistently, pDCs lacking Atg5 failed to secrete IFN-α and IL-12p40 in response to VSV infection, in a previous study. In addition to VSV, a TLR9 ligand herpes simplex virus (HSV)-1 failed to induce IFN-α production in Atg5-deficient pDCs, while the IL-12 response remained intact in these cells.31 These data suggest that the IFN-α signaling pathway is impaired in the absence of Atg5. However, the precise nature of the differential control of NF-κB versus IFN-α induction pathways in pDCs by autophagy remains to be determined.33
Although TLRs are the most well-known PRRs, some bacterial structures are recognized by intracellular sensors, called nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs).34,35 NLRs are a family of cytoplasmic molecules that recognize bacterial cell wall components, such as peptidoglycans, and initiate signaling cascades to activate inflammatory responses.35 NLRs comprise two different subsets, NODs and NALP (NACHT-, LRR- and pyrin domain-containing proteins). When NLRs recognize bacterial peptidoglycans, they initiate signaling pathways by recruiting protein kinase, which in turn activates NF-κB and AP-1, leading to production of cytokines and other molecules of innate immunity.36 Recent studies have shown that stimulation of NOD2 by muramyl dipeptide induces activation of autophagy and clearance of bacteria.37 This effect depends on autophagy-related proteins, such as Atg5, Atg7, and Atg16L1, as well as receptor interacting serine-threonine kinase-2, a regulator of the NOD2 signaling pathway. When NOD2 is stimulated with bacterial components, the autophagy protein Atg16L1 is recruited to the plasma membrane at the bacterial entry site.38 This then promotes bacterial trafficking to the autophagosome, maturation of the autophagosome via fusion with a lysosome, and facilitates antigen loading to MHC class II molecules to enhance antigen presentation to CD4 T cells. As expected, DCs isolated from patients of Crohn's disease, expressing risk alleles for NOD2 or Atg16L1, are not able to induce autophagy pathways, and in turn, fail to promote bacterial clearance and antigen presentation.37,38
Most cell types utilize cytoplasmic receptors, RIG-I-like receptors (RLRs), to bind to viral RNA. These cytosolic sensors are caspase recruitment domain (CARD)-containing proteins, consisted of retinoic acid inducible gene I (RIG-I) and melanoma differentiation associated gene 5.39-41 Via their CARD domain, RLRs signal with IFN-β promoter stimulator-1 (IPS-1) and activate transcription factors, including IRF3 and NF-κB, to produce type I IFN and proinflammatory cytokines, respectively.36 Although autophagy-related proteins such as Atg5 mediate transport of viral replication intermediates from the cytosol to lysosomes, where recognition thereof by TLR7 occurrs,31 autophagy-related proteins engaged in RLR-mediated viral sensing act as negative regulators of anti-viral responses.42 The Atg5-Agt12 conjugate directly associates with CARD domains of RIG-I and IPS-1 to suppress type I IFN production (Fig. 1B). Consequently in Atg5-deficient mouse embryonic fibroblasts (MEFs), RLR signaling resulted in the overproduction of type I IFNs and resistance to VSV infection.42 Moreover, the accumulation of dysfunctional mitochondria and ROSs, associated with dysfunctional mitochondria in Atg5-deficient cells, potentiated RLR signaling in response to viral infections.43
In cases where double-stranded DNA (dsDNA), derived from bacteria or DNA viruses, is detected, the sensing and regulation mechanisms thereof are still unclear. Recently, studies have shown that dsDNA can induce the production of type I IFNs.44,45 In this process, the translocation and assembly of stimulator of IFN genes (STING) and TANK-binding kinase 1 (TBK1) are required.46-49 After sensing dsDNA, STING, a multispanning membrane protein, is translocated from the ER to the Golgi apparatus and assembled with TBK1, which phosphorylates the transcription factor IRF3, resulting in the production of type I IFN. Atg9a, an essential component of autophagy, is co-localized with STING in the Golgi apparatus and controls the dynamic translocation of STING and assembly with TBK1 (Fig. 1B).50 In Atg9a-deficient MEFs, but not in Atg7-and Atg16L1-deficient MEFs, the translocation of STING from the Golgi apparatus to the cytoplasmic punctate structures and assembly with TBK1 are greatly enhanced, which, in turn, induces aberrant activation of type I IFN responses.25 Thus, these results demonstrated that Atg9a plays a role in the fine-tuning of innate immune responses.
Autophagy has been known to be involved in innate immunity by directly defending against pathogens and by promoting innate recognition of pathogens. Furthermore, autophagy facilitates adaptive immune responses such as antigen presentation.51,52 Generally, intracellular cytosolic or nuclear antigens are presented by MHC class I molecules to CD8 T cells after proteosomal hydrolysis, whereas extracellular antigens are presented by MHC class II molecules to CD4 T cells after lysosomal degradation.53,54 Thus, it is somewhat expectable that lysosomal degradation of cytoplasmic molecules by autophagy can provide antigens presented by MHC class II molecules to activate CD4 T cells (Fig. 1D). Indeed, cytosolic and nuclear antigens are composed of 20-30% of natural ligands of MHC class II molecules.55-57 Recent studies revealed that autophagy is involved in the MHC class II processing and presentation of various intracellular antigens to CD4 T cells.17-21 Macrophages, which are unable to present several epitopes of endogenous C5 protein (5th component of mouse complement) with MHC class II molecules, became able to do so when they were treated with low doses of the lysosomotropic agent ammonium chloride.17 Also, MHC class II presentation was abrogated when treated with inhibitors of the class III PI3 kinase, which is known to inhibit autophagy induction. The presentation of peptides from the tumor antigen mucin 1 with transfection into DCs by RNA electroporation was inhibited to present by treatment of the inhibitor of class III PI3 kinase, 3-methyladenine (3-MA) and Wortmannin.18 Endogenous presentation of cytosolic protein neomycin phosphotransferase II (NeoR) on MHC class II was mediated by autophagy. NeoR was sequestrated into autophagosomes and delivered to lysosomes. This pathway was blocked by 3-MA and Wortmannin.19 Another report indicated that the endogenous nuclear antigen 1 of EBV (EBNA1) gains access to the MHC class II presentation pathway via autophagy. EBNA1 was accumulated in autophagosomes when lysosomal acidification was inhibited. Further, intracellular EBNA1 processing for MHC class II presentation was inhibited after siRNA-mediated silencing of Atg12, an essential protein involved in autophagosome formation.20 A more recent report showed that the targeting of a model antigen, influenza matrix protein 1 (MP1), to autophagosomes enhanced MHC class II presentation.21 Via fusion of MP1 to LC3, an autophagosome-associated protein, antigen presentation with MHC class II to MP1-specific CD4 T cells was enhanced 20-fold, while MHC class I presentation to CD8 T cells was not affected. Furthermore, siRNA-mediated silencing of Atg12 inhibited MP1-LC3 transport to the MHC class II compartment. This result implicated that autophagy plays important roles in delivering cytosolic antigens to the MHC class II compartment for enhanced MHC class II presentation to CD4 T cells.
In recent years, studies have revealed that autophagy promotes MHC class II presentation of extracellular antigens to CD4 T cells.58,59 In mice with dendritic cell (DC)-conditional deletion in Atg5, CD4 T cell priming after herpes simplex virus infection was impaired.58 Atg5-deficient DCs showed a pronounced defect in the fusion of phagosomes with lysosomes and consequent processing and presentation of phagocytosed antigens containing TLR stimuli for MHC class II. Similarly, autophagy mediated engulfment of endocytosed extracellular human immunodeficiency virus-1 (HIV-1) and promoted the processing of peptides for presentation to MHC class II loading compartments.59 In this report, envelope proteins of HIV-1 downregulated autophagy pathways by enhancing mammalian target of rapamycin signaling, and consequently autophagy failed to promote MHC class II presentation.
The involvement of autophagy in immunity suggests that autophagy may be a target for therapy in various human diseases. For example, antigens from pathogens, which induce severe infectious diseases and are targeted by autophagy pathways, can be used to develop vaccines in order to enhance MHC class II presentation to CD4 T cells. Also, autophagy may be developed to antimicrobial agents based on the ideas that enhancing or inhibiting autophagy affects host resistance to viral, bacterial and parasitic infection. Furthermore, as mutations of autophagy genes such as Atg16L1 are known to be causal factors of Crohn's disease,37,38 modulation of autophagy might ameliorate Crohn's disease, if it can compensate for the functions of Atg16L1. The basic function of autophagy, degrading damaged or long-lived organelles might be a useful target for cancer therapy. However, controversy remains on whether enhancing or inhibiting autophagy might have beneficial effects on cancer therapy.60-63 In addition, imbalanced control of autophagy has demonstrated adverse effects including disturbed negative selection with consequent autoimmunity.64,65 Therefore, further comprehensive understanding of autophagy will be needed for proper modulation thereof without unwanted immunologic consequences.
Recent studies have demonstrated that autophagy is important to immune responses. With its most notable function of degrading cytosolic constituents, it has also been shown to degrade intracellular microbes. In addition to directly defending against pathogens, autophagy facilitates the sensing of pathogens by pattern recognition receptors and modulates consequent signaling pathways in multiple ways. Autophagy promotes the elimination of some bacteria by autophagosome formation, whereas some fungal infections are resolved through maturation of phagosomes with the assistance of autophagy. Moreover, autophagy promotes the audetection of viral infections by delivering the cytosolic replication complex of the virus to the site of endosomal sensor recognition. Although autophagy usually enhances the signaling pathway after PRRs sense the pathogens, it can also negatively regulate the signaling pathway in certain cases. In addition to innate recognition, autophagy can be used to facilitate MHC class II presentation of endogenous intracellular antigens as well as phagocytosed or endocytosed extracellular antigens to activate CD4 T cells. Since autophagy plays multiple roles in immunity, autophagy is a potential target for therapies in various human diseases. However, it is difficult to determine whether induction or suppression of autophagy has beneficial effects, since autophagy is involved in multiple pathways of immune regulation. Thus, further comprehensive understanding of autophagy will provide a more integrated picture of how this system controls innate and adaptive immunity.
Figures and Tables
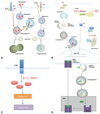 | Fig. 1Autophagy contributes to innate and adaptive immune responses against pathogens. (A) TLR promotes the induction of autophagy for pathogen elimination. TLR4 signaling via the TRIF-p38 axis, but not via MyD88, induces the formation of autophagosome and subsequent elimination of Mycobacteria bacilli. TLR7 signaling induced by two different ligands, single-stranded RNA and imiquimod, induces the formation of an autophagosome to eliminate Bacillus Calmette-Guerin. Atgs, such as Atg5, beclin and PI3K, are required for the formation of autophagosomes via TLR stimulation. In another case, Atg facilitates the TLR-dependent elimination of pathogens by promoting phagosome maturation. When zymosan (a particle of fungal cell walls) is phagocytosed, TLR2 induces the recruitment of LC3 to the phagosomal membrane, leading to the maturation of phagosomes and elimination of the fungus. This process depends on Atg5 and Atg7. (B) Autophagy supports antiviral responses by promoting viral sensing. In pDCs, autophagy mediates the recognition of the virus infection by delivering viral replication intermediates in the cytosol to lysosomes, where TLR recognition occurs. However, in non-pDCs, autophagy negatively regulates antiviral type I IFN responses. The Atg5-Agt12 conjugate directly associates with CARD domains of RIG-I, a cytosolic receptor for viral nucleic acids, and IPS-1, suppressing type I IFN production. Consequently in Atg5-deficient MEFs, RLR signaling results in overproduction of type I IFNs and resistance to VSV infection. In cases of dsDNA recognition, STING, a multispanning membrane protein, is translocated from the ER to the Golgi apparatus and assembled with TBK1, which phosphorylates the transcription factor IRF3, resulting in the production of type I IFN. Atg9a, an essential component of autophagy, is co-localized with STING in the Golgi apparatus and controls the dynamic translocation of STING and assembly with TBK1. In Atg9a-deficient MEFs, the translocation of STING from the Golgi apparatus to the cytoplasmic punctate structures and assembly with TBK1 are greatly enhanced, which, in turn, induces aberrant activation of type I IFN responses. (C) Atg16L1 regulates endotoxin-induced inflammasome activation. In autophagy-deficient cells, IL-1β and IL-18 production is enhanced in response to LPS. Macrophages lacking Atg16L1 induce high-levels of ROS, which, in turn, activates caspase-1, leading to the processing of IL-1β. However, in macrophages of wild-type mice, generation of ROS is inhibited by autophagy-related protein, leading to limited production of IL-1β. (D) Autophagy is involved in MHC class II processing and presentation of intracellular antigens to CD4 T cells. Intracellular antigens are engulfed by autophagosomes, transported to MHC class II loading compartments (MIIC), and then loaded onto MHC class II molecules for presentation to CD4 T cells. TLR, toll-like receptor; Atg, autophagy-related genes; pDCs, plasmacytoid dendritic cells; CARD, caspase recruitment domain; RIG-I, retinoic acid inducible gene I; MEFs, mouse embryonic fibroblasts; RLR, RIG-I-like receptor; VSV, vesicular stomatitis virus; STING, stimulator of IFN genes; TBK1, TANK-binding kinase 1; LPS, lipopolysaccharides; ROS, reactive oxygen species; TRIF, TIR domain-containing adapter-inducing interferon-β; IFN, interferon; IPS-1, IFN-β promoter stimulator-1; ER, endoplasmic reticulum; IRF, interferon regulatory factor; IL, interleukin; MHC, major histocompatibility complex. |
ACKNOWLEDGEMENTS
This study was supported by the Korean Health Technology R&D Project (A100920) and the National R&D Program for Cancer Control (1020230) funded by the Ministry of Health & Welfare, Republic of Korea. This study was also supported by the National Research Foundation (2010-0012891) and the Converging Research Center Program (2011K000864) funded by the Ministry of Education, Science and Technology of Korea.
References
1. Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000. 290:1717–1721.

2. Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004. 432:1032–1036.

3. Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005. 169:425–434.

4. Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007. 27:19–40.

5. Massey AC, Zhang C, Cuervo AM. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol. 2006. 73:205–235.

6. Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci. 2000. 113(Pt 24):4441–4450.

7. Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996. 273:501–503.

8. Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989. 246:382–385.

9. Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, et al. A protein conjugation system essential for autophagy. Nature. 1998. 395:395–398.

10. Klionsky DJ, Cregg JM, Dunn WA Jr, Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003. 5:539–545.

11. Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002. 27:421–429.

12. Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004. 6:463–477.
13. Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004. 306:990–995.

15. Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol. 2004. 2:301–314.

16. Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005. 120:159–162.

17. Brazil MI, Weiss S, Stockinger B. Excessive degradation of intracellular protein in macrophages prevents presentation in the context of major histocompatibility complex class II molecules. Eur J Immunol. 1997. 27:1506–1514.

18. Dörfel D, Appel S, Grünebach F, Weck MM, Müller MR, Heine A, et al. Processing and presentation of HLA class I and II epitopes by dendritic cells after transfection with in vitro-transcribed MUC1 RNA. Blood. 2005. 105:3199–3205.

19. Nimmerjahn F, Milosevic S, Behrends U, Jaffee EM, Pardoll DM, Bornkamm GW, et al. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol. 2003. 33:1250–1259.

20. Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, et al. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005. 307:593–596.

21. Schmid D, Pypaert M, Münz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007. 26:79–92.

23. Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004. 5:987–995.

25. Saitoh T, Akira S. Regulation of innate immune responses by autophagy-related proteins. J Cell Biol. 2010. 189:925–935.

26. Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007. 27:135–144.

27. Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008. 456:264–268.

28. Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008. 27:1110–1121.

29. Delgado MA, Deretic V. Toll-like receptors in control of immunological autophagy. Cell Death Differ. 2009. 16:976–983.

30. Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, et al. Toll-like receptor signalling in macrophages links the audetection tophagy pathway to phagocytosis. Nature. 2007. 450:1253–1257.

31. Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007. 315:1398–1401.

34. Inohara , Chamaillard , McDonald C, Nuñez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005. 74:355–383.

35. Martinon F, Tschopp J. NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 2005. 26:447–454.

36. Lee MS, Kim YJ. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu Rev Biochem. 2007. 76:447–480.

37. Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010. 16:90–97.

38. Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhães JG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010. 11:55–62.

39. Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004. 5:730–737.

40. Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005. 175:2851–2858.

41. Foy E, Li K, Sumpter R Jr, Loo YM, Johnson CL, Wang C, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A. 2005. 102:2986–2991.

42. Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007. 104:14050–14055.

43. Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A. 2009. 106:2770–2775.

44. Charrel-Dennis M, Latz E, Halmen KA, Trieu-Cuot P, Fitzgerald KA, Kasper DL, et al. TLR-independent type I interferon induction in response to an extracellular bacterial pathogen via intracellular recognition of its DNA. Cell Host Microbe. 2008. 4:543–554.

45. Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006. 24:93–103.

46. Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008. 455:674–678.

47. Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008. 28:5014–5026.

48. Sun W, Li Y, Chen L, Chen H, You F, Zhou X, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009. 106:8653–8658.

49. Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008. 29:538–550.

50. Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009. 106:20842–20846.

53. Sijts EJ, Kloetzel PM. The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell Mol Life Sci. 2011. 68:1491–1502.

54. van den Hoorn T, Paul P, Jongsma ML, Neefjes J. Routes to manipulate MHC class II antigen presentation. Curr Opin Immunol. 2011. 23:88–95.

55. Marrack P, Ignatowicz L, Kappler JW, Boymel J, Freed JH. Comparison of peptides bound to spleen and thymus class II. J Exp Med. 1993. 178:2173–2183.

56. Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999. 50:213–219.

57. Muntasell A, Carrascal M, Serradell L, Veelen Pv P, Verreck F, Koning F, et al. HLA-DR4 molecules in neuroendocrine epithelial cells associate to a heterogeneous repertoire of cytoplasmic and surface self peptides. J Immunol. 2002. 169:5052–5060.

58. Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010. 32:227–239.

59. Blanchet FP, Moris A, Nikolic DS, Lehmann M, Cardinaud S, Stalder R, et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010. 32:654–669.

60. Rosenfeldt MT, Ryan KM. The role of autophagy in tumour development and cancer therapy. Expert Rev Mol Med. 2009. 11:e36.

61. Levine B. Cell biology: autophagy and cancer. Nature. 2007. 446:745–747.
62. Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007. 7:961–967.

63. Wilkinson S, Ryan KM. Autophagy: an adaptable modifier of tumourigenesis. Curr Opin Genet Dev. 2010. 20:57–64.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download