INTRODUCTION
The increasing prevalence of antimicrobial resistant bacteria has become a serious worldwide problem. The prevalence of antibiotic-resistant bacteria varies greatly from country to country, because it is greatly influenced by antimicrobial usage and failure to control the spread of resistant bacteria. Antimicrobial-resistant bacteria have been relatively more prevalent in Asian countries
1 and in some European countries such as Spain, Italy and France (
www.ecdc.europa.eu). Although bacterial resistance has generally been rare in Scandinavian countries, rare antimicrobial-organism combinations are being reported with increasing frequency, mostly due to importation from other countries.
2
In general, resistant bacteria have been less prevalent in the U.S. and in Europe; however, vancomycin-resistant enterococci were first reported in the U.K. and France in 1988.
3,
4 Following the first discovery of highly vancomycin-resistant VanA-producing
Staphylococcus aureus in the U.S., nine more isolates were subsequently detected, while only one of each was isolated in India and Iran.
5 The KPC type carbapenemase-producing
Klebsiella pneumoniae emerged in the U.S.
6 and spread to other countries.
Resistance of Gram-positive cocci was once considered to be a more serious problem, but the emergence of carbapenem-resistant Gram-negative bacilli, either due to class A β-lactamase, KPC,
6 or class B β-lactamases, IMP, VIM, recently identified NDM-1, etc.
7,
8 raised great concern once again about the resistance of Gram-negative bacilli, because these enzymes can inactivate the most potent class of β-lactam antibiotics, i.e. carbapenems.
An antimicrobial resistance surveillance study is a fundamental means of appreciating trends in resistance, developing accurate treatment guidelines, and evaluating the efficacy of intervention (
www.who.int/drugresistance/en/). Monitoring the temporal trend of resistance is an essential element of the detection of subtle variations in antimicrobial resistance.
9 The Korean Nationwide Surveillance of Antimicrobial Resistance (KONSAR) program has been conducted for the purpose of surveillance since 1997.
10 The study from 2007
11 showed continued prevalence of methicillin-resistant
S. aureus (MRSA), third-generation cephalosporin-resistant
K. pneumoniae, and fluoroquinolone-resistant
Escherichia coli,
Pseudomonas aeruginosa and
Acinetobacter spp. A high amikacin resistance rate in
Acinetobacter spp., and, more importantly, a gradual increase in vancomycin-resistant
Enterococcus faecium and imipenem-resistant
Acinetobacter spp., was also found. In addition, third-generation cephalosporin-resistant
E. coli and
K. pneumoniae, and imipenem-resistant
P. aeruginosa were highly prevalent among the commercial laboratory (Com Lab) tested isolates.
In the present study, the trends of resistance of the aforementioned antimicrobial-organism combinations were determined. Stenotrophomonas maltophilia was also added to the surveillance study, because this organism is naturally resistant to quite a few classes of antimicrobial agents, and because it has been increasingly detected. Because intensive care units (ICUs) are a major source of resistant bacteria, resistance rates of isolates from ICUs and non-ICUs were compared using the data generated at six large hospitals with >1,000 beds. Resistance rates of isolates from secondary-care hospitals and from primary care clinics were also compared using data from a Com Lab.
DISCUSSION
The prevalence of resistant organisms varies significantly by time, country, and patient population. Therefore, antimicrobial resistance surveillance studies are very important to monitor levels of endemic or emerging resistance. In fact, with rapid increases in resistant bacteria, we need to devote more surveillance efforts to help delay the increasing trend of resistance, although it is impossible to completely overcome the problem. An antimicrobial surveillance study is done either by testing collected isolates at a coordinating laboratory or by analyzing the susceptibility data generated by each hospital laboratory. It was considered that the present need is for a supranational resistance surveillance system including non-invasive isolates, preferably based on all routine susceptibility data obtained from local laboratories.
19
The present KONSAR study showed that the majority of hospital laboratories used commercial broth microdilution methods for susceptibility testing of E. coli and S. aureus, which can represent Gram-negative and Gram-positive bacteria, respectively. Automation and rapid results are some of the advantage of these methods, but they may not perform optimally for the detection of some antimicrobial-organism carbapencombinations. With the increasing prevalence of multidrug resistant (MDR) Acinetobacter spp. and P. aeruginosa, polymyxins and tigecycline susceptibility testing have become a requirement, but these were not included in the present study mainly because of their unavailability in the majority of commercial systems.
In 2010, the CLSI
13 lowered the resistance breakpoints of cefotaxime, ceftazidime, and aztreonam to ≥4 µg/mL, ≥16 µg/mL, and ≥16 µg/mL, respectively, and eliminated the necessity of ESBL testing of
E. coli,
K. pneumoniae spp., and
P. mirabilis. In our present study, the previous CLSI breakpoints
12 were used for these antimicrobial agents, and some laboratories differentiated ESBL-producing isolates.
In the analysis of laboratory generated susceptibility data, the CLSI
20 recommends including only the first isolate if there were multiple isolates from one patient, for the purpose of guiding clinicians in the empirical selection of appropriate antimicrobial agents. The WHONET software
14 can be used to exclude duplicate isolates. In the present study, the Com Labs were not able to exclude duplicate isolates due to difficulty identifying relevant patients. This apparently raised the resistance rates of some organisms to certain antimicrobial agents. However, if only the first isolate is included, and particularly if the analysis is made only once a year, it may underestimate resistance acquired during hospitalization, as well as the emergence of resistance due to mutations during therapy.
19 A study showed that not all U.S. hospitals excluded duplicate isolates.
21 In fact, to detect developing resistance during therapy, the CLSI
13 recommends testing the susceptibility of subsequent isolates three to four days after the initial isolation of
Enterobacter,
Citrobacter, and
Serratia spp. to third-generation cephalosporins, of
P. aeruginosa to all antimicrobial agents and of staphylococci to quinolones.
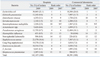
In 2009, as in 2007,
E. coli,
S. aureus, CNS,
P. aeruginosa, and
K. pneumoniae were the five most prevalent organisms among those isolated by hospitals (
Table 1). Among the Com Lab isolates,
E. coli,
P. aeruginosa, and
E. faecium were ranked numbers 1, 2, and 5, respectively, in both 2007 and 2009, but the rank orders of
S. aureus and
K. pneumoniae were reversed (3 and 4 in 2007; 4 and 3 in 2009, respectively). The rank orders in the U.S. were largely similar. The SENTRY Program in 2004-2008 showed that the five most frequently isolated bacteria from hospitalized patients with pneumonia were
S. aureus (36.3%),
P. aeruginosa (19.7%),
Klebsiella spp. (8.5%),
E. coli (4.6%) and
Acinetobacter spp. (4.8%). The proportion of
S. maltophilia was 3.3% and rank order was No. 8.
22 In our study, the rank order of S. maltophilia was similar, i.e., No. 10.
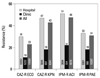
Our present Com Lab data showed that isolates from secondary-care hospitals were much more frequently resistant than those from primary-care clinics, suggesting a higher prevalence of nosocomially acquired pathogens among the hospital isolates (
Fig. 6). In Germany, hospitals with their own microbiology laboratory reported higher frequencies of health care-associated infections than did hospitals without in-house laboratory services.
23
In a recent review, Peleg and Hooper
24 expressed concern about hospital-acquired Gram-negative bacterial infections. In the U.S., it was estimated that a total of 1.7 million hospital-acquired infections occurred (4.5 per 100 admissions) in 2002, and hospital-acquired infections were the sixth leading cause of death. It is of great concern that only approximately one third of hospital-acquired infections are preventable.
ICUs are reservoirs of resistant bacteria. The U.S. National Healthcare Safety Network (NHSN) indicated that Gram-negative bacteria caused more than 30% of hospital-acquired infections, and these bacteria predominated in cases of ventilator-associated pneumonia (47%) and urinary tract infections (45%). In ICUs, Gram-negative bacteria accounted for about 70% of these types of infections.
24 An international study showed that resistance rates of ICU isolates from Latin America, Asia, Africa, and Europe were higher than those in the U.S.: MRSA 84.1% vs. 56.8%,
K. pneumoniae to ceftazidime or ceftriaxone 76.1% vs. 27.1%,
A. baumannii to imipenem 46.3% vs. 29.2%, and
P. aeruginosa to piperacillin 78.0% vs. 20.2%, respectively.
25 However, it is noteworthy that in the U.S., the proportion of MRSA was high, and those of other organisms were not low.
Resistance is reported to have increased throughout Europe, among Gram-negative bacilli in particular.
19 Gram-negative bacterial infections have features that are of particular concern. These organisms are highly efficient at up-regulating or acquiring resistance genes.
24 In our present study, the ceftazidime resistance rate of hospital isolates of
K. pneumoniae was 33%. The rate was much higher than the 0% among 27 isolates in Iceland, but much lower than the 68.7% found in Greece (
www.ecdc.europa.eu). Among the Gram-negative bacilli associated with intra-abdominal infections in 2008 from 11 Asia-Pacific countries, the highest rates of ESBL-producing
E. coli and
K. pneumoniae were observed in India (61.2% and 46.8%, respectively); the rates were 30.4% and 25.0% in Korea, respectively.
1
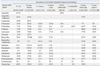
Currently the most feared resistance is those to carbapenems. In our present study, 26% of
P. aeruginosa were resistant to imipenem (
Table 2,
Fig. 4). This rate is much higher than the 0% of 16 isolates found in Iceland, but much lower than the 44% found in Greece (
www.ecdc.europa.eu). Of over 800 β-lactamases identified from Gram-negative bacilli, at least 120 have been detected in
P. aeruginosa. IMP- and VIM-type MBLs are predominantly found in
P. aeruginosa and in
Acinetobacter spp.
26 The proportions of imipenem-resistant isolates in the 1997 vs. 2009 KONSAR studies were:
P. aeruginosa 17% vs. 26%, and
Acinetobacter spp. 1% vs. 51%, respectively (
Figs. 3 and
4). The drastic increase in imipenem-resistant
Acinetobacter spp. was probably due to OXA-type carbapenemase production. It is interesting that almost all carbapenem-resistant
A. baumannii carried
blaOXA-23-like or ISA
ba1-activated
blaOXA-51-like genes, while the majority of non-
baumannii Acinetobacter carried MBL genes.
27
The terms MDR, extreme drug resistance (XDR), and pandrug resistance (PDR), are very useful to attract attention, but there has been no consensus as to their definition. The term PDR in
A. baumannii and
P. aeruginosa in particular is thought to have been used inappropriately.
28 In one study, MDR was defined as nonsusceptibility to one or more antimicrobials in three or more antimicrobial classes (antipseudomonal penicillins, third-generation cephalosporins, carbapenems, fluoroquinolones, and aminoglycosides). The definition did not include colistin and tigecycline susceptibility.
29 The study showed that the proportions of MDR isolates of
A. baumannii and
P. aeruginosa reported to the U.S. NHSN systems in 2008 were 74% and 17%, respectively, but XDR isolates (nonsusceptible to all antimicrobials including polymyxins and tigecycline) remained rare. Of the 344
blaKPC-positive ICU isolates of Enterobacteriaceae, 91% and 99% were susceptible to colistin and tigecycline, respectively. Only two isolates were nonsusceptible to both antimicrobial agents. In Korea, colistin-resistant
A. baumannii isolates were detected.
30 Therefore, it is necessary to include colistin and tigecycline susceptibility in future surveillance studies in Korea.
In our present study, 44% of hospital isolates of
H. influenzae were resistant to ampicillin. Bae, et al.
31 reported that among 544 nationwide collections of respiratory isolates of
H. influenzae, 58.5% were ampicillin resistant, and 52.4% were β-lactamase producers.
In Korea, hospital-associated MRSA has been a serious problem (
Fig. 1). MRSA has become a problem organism in North America as well. The SENTRY Program showed that among the
S. aureus isolates in 2008 from hospital-acquired and ventilator-associated bacterial pneumonia in North America, 54% were nonsusceptible to oxacillin.
22 Vancomycin-resistant enterococci are also important Gram-positive nosocomial pathogens. In the U.S., hospitalizations with vancomycin-resistant enterococcal infections increased from 3.16 to 6.51 per 10,000 hospitalizations during 2003-2006.
32 The U.S. NHSN data
33 showed that 80% of
E. faecium isolates in 2006 and 2007 from central line-associated bloodstream infections, catheter-associated urinary tract infections, ventilator-associated pneumonia, and surgical site infections were vancomycin resistant.
The EARS-Net 2009 report (
www.ecdc.europa.eu) showed that the prevalence of invasive MRSA and vancomycin-resistant
E. faecium varied greatly by country. For example, the proportions of MRSA were 0% of 59 isolates in Iceland, and 49.1% of 1,824 isolates in Portugal, and those of vancomycin-resistant
E. faecium were 0% of 243 isolates in Finland, and 37.8% of 386 isolates in Ireland. The report also showed that the resistance rates varied greatly depending on hospitals even within a country: the mean prevalence of MRSA from Germany was 18.7%, but ranged from a minimum of 0% to a maximum of 60%. In our present study, 39% of hospital isolates of
S. pneumoniae were resistant to penicillin G at the CLSI meningitis breakpoint.
13 This rate did not differ greatly from those of a U.S. study, 28.4% in 1998 and 41.0% in 2009.
34 Until 2008, penicillin G susceptibility of all
S. pneumoniae isolates was interpreted according to CLSI breakpoint for meningitis, because a non-meningitis breakpoint did not exist. In our present report, we continued to include oxacillin screening test results for respiratory isolates, because the majority of
S. pneumoniae serotypes recovered from the respiratory tract are those causing meningeal infections, too, and because the result could show a resistance trend.
With increasing international air travel and import of animal produce, it has become very difficult to control the spread of antimicrobial-resistant bacteria. Carbapenemases IMP-1 and VIM-2, initially detected in Japan
35 and in France,
7 respectively, are now widespread globally, including in Korea. Scandinavia is a region with a low prevalence of antimicrobial resistance, but import and local clonal expansion of VIM-2-producing
P. aeruginosa has been reported.
2 Also,
blaSPM-1, which was initially found in Brazil, was detected recently in a
P. aeruginosa isolate in Switzerland.
36 An isolate of
K. pneumoniae, producing a new MBL, NDM-1, was first found in Sweden in a patient transferred from India.
8 K. pneumoniae isolates producing a class A carbapenemase, KPC-2, were first found in New York.
6 K. pneumoniae Bidisolates, carrying NDM-1 (unpublished information), and carrying KPC-2
37 were detected in Korean patients in 2010.
We now know that use of antimicrobial agents is the cause of the emergence and spread of resistant bacteria. However, we should acknowledge that antimicrobial resistant bacteria existed before the clinical use of antimicrobial agents. We also should know that stopping the prescription of antimicrobial agents to outpatients with colds may not reduce the emergence of certain resistances such as to vancomycin and carbapenems. It has been hoped that resistant bacteria could be reduced by stopping overuse of antimicrobial agents. However, it is difficult to judge the overuse of antimicrobial agents at a hospital or ward. A study in France in 2007 showed the level of antimicrobial consumption to be only 60 defined daily doses (DDD)/1,000 patient-days (PD) in psychiatric wards compared to 1,466 DDD/1,000 PD in ICUs. Glycopeptides and carbapenems were mostly used in cancer patients and in teaching hospitals.
38 Sykes
39 stated that however hard we try and however clever we are, bacteria that have adapted to survive under the most extreme conditions for the past three billion years, and will overcome whatever we do to control them. The emergence of antibiotic resistance is inevitable, but we must seek to decrease its impact and to prolong the effectiveness of the agents available to us.
40 For this purpose, continued surveillance study of antimicrobial resistance is needed more than ever before to determine the trends of already prevalent resistant organisms and to detect the recently imported KPC-2 and NDM-1-producing Gram-negative bacilli.
In conclusion, the present surveillance study in 2009 showed that among hospital laboratory isolated bacteria, the prevalence of ceftazidime-resistant K. pneumoniae, fluoroquinolone-resistant K. pneumoniae, Acinetobacter spp., and P. aeruginosa further increased, and imipenem-resistant Acinetobacter spp. increased drastically. It was also noted that methicillin-resistant S. aureus remained highly prevalent, and vancomycin-resistant E. faecium further increased. Problematic antimicrobial-organism combinations were much more prevalent among ICU isolates, and were also prevalent among isolates tested at Com Labs.









 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download