Abstract
Purpose
Surgical therapy is the primary treatment for oral cancer, but it can cause facial distortion. Therefore, if anticancer drugs are effective against oral cancer, they may be used preferentially. However, oral squamous carcinoma cells (OSCCs) are resistant to these drugs, so finding a way to enhance the sensitivity of these cells to anticancer drugs is important. The bacterial protein azurin is known to selectively enter cancer cells and induce apoptosis. In this study, we show the anticancer effect of azurin in OSCC.
Materials and Methods
OSCC cell line (YD-9) was subjected to azurin treatment. Cell viability, morphology and protein expression levels were monitored after treatment of azurin. Cells were also subjected to combination treatment of azurin with either 5-fluorouracil or etopside.
Results
Azurin-treated cells showed decreased cell viability accompanied by apoptotic phenotypes including morphological change, DNA breakage, and increases in p53 and cyclin B1 protein levels. Combination treatment of azurin with other anti-tumor agents caused an increase in sensitivity to anticancer drugs in azurin-treated YD-9 cells.
Oral cancer is one of the 10 most frequently occurring cancers worldwide,1 with more than 90% of malignancies being oral squamous cell carcinoma (OSCC), which originates from the oral mucosa. OSCC resistance to drugs may be circumvented by combining drugs, because the drugs can activate alternate apoptotic pathways when used in combination. Therefore, the clinical application of a combination of drugs may have synergistic effects, reducing resistance and/or cytotoxicity.2
Azurin, a cupredoxin type of electron transfer and purified low molecular weight redox protein from the pathogenic bacteria Pseudomonas aeruginosa, selectively induces and triggers apoptosis in certain human cancer cells. Recently, Yamada, et al.3 reported that azurin can effectively enter human cancer cells but not normal cells. After internalization, azurin forms a complex with the tumor suppressor protein p53 and stabilizes it,4 thereby inducing apoptosis or cell cycle arrest in the G1 phase.5,6
Despite extensive analysis of azurin's antitumor activity, its ability to modulate oral cancer growth has not yet been characterized. In this study, the antitumor effect of azurin on YD-9 and MG-63 cells is elucidated. Azurin controls p53 and cyclin B1 protein levels, leading to apoptosis of OSCC. Furthermore, combination treatment of azurin with 5-fluorouracil or etoposide effectively increases the sensitivity of OSCC to anticancer drugs.
The azurin gene was amplified using PCR with genomic DNA of the P. aeruginosa strain as the template DNA. Primer sequences were 5'-GCCCAAGCTTACCTAGGAGGCTGCTCCATGCTA-3' and 5'-TGAGCCCCTGCAGGCGCCCATGAAAAAGCCCGGC-3'; the additionally introduced HindIII and PstI sites are underlined. A plasmid expressing GST-tagged azurin was constructed by introducing it into pGEX-4T-2. To purify the azurin protein, BL21 (DE3) cells were used as the host strain. After induction with 1 mM IPTG, GST-fusion proteins were purified from cell extracts, with affinity chromatography using an ÄKTA FPLC system (Amersham Biosciences, Little Chalfont, UK). Next, 5 µg of purified protein was used for 15% SDS-PAGE and stained with staining buffer containing Coomassie brilliant blue.
The YD-9 cell line established from an OSCC patient (obtained from the Korean Cell Line Bank) and the human osteosarcoma cell line MG-63 (ATCC, CRL-1427, HTB 96, Manassas, VA, USA) were used in this study. The cells were cultured in Dulbecco's modified Eagle medium (Gibco, NY, USA) with nutrient mixture F12 (DMEM/F12, 3:1) supplemented with 10% heat-inactivated fetal bovine serum (FBS, GIBCO), 1% glutamine and 100 µg/mL penicillin/streptomycin at 37℃ in a humidified incubator with 5% CO2 atmosphere.
To elucidate the effects of purified azurin combined with anticancer drugs, YD-9 and MG-63 cells were cultured in a 96-well plate. After 24 hours of incubation, the medium was replaced with fresh medium containing various concentrations of 5-fluorouracil (5-FU, Choongwae Pharma Corporation, Seoul, Korea) or etoposide (Sigma, St. Louis, MO, USA) along with 200 µg/mL azurin.
The cells were suspended in a 96-well plate (SPL Life Science, Pocheon, Korea) at a density of 2×104 cells per well. After 24 h, they were treated with various concentrations of azurin, 5-FU, or etoposide for different periods of time (0-72 h). Next, the cells were treated with 1 mg/mL MTT in the growth medium for 4 h at 37℃. Cell viability was evaluated by comparison with a control culture (assumed to be 100% viable), measuring the intensity of the blue color (OD at 570 nm) using a multi-well reader (Quant, Bio-Tek, Highland Park, VT, USA).
The cells were washed with cold PBS and incubated with 4 µg/mL of Hoechst 33342, a DNA-binding fluorescent dye, for 30 min at 37℃ in the dark. Then, the cells were fixed using 4% paraformaldehyde for 10 min. The morphological characteristics of the apoptotic cells were identified under a fluorescent microscope (ECLIPSE E800, Nikon, Tokyo, Japan) with excitation at 540 nm.
The cells were prepared on microscope slides, air dried, and then immersed in a fixative solution. These slides were immersed in color reagent red and blue solution. The cells were washed twice with PBS and mounted in 50% glycerol in PBS. The morphological characteristics of the cells were examined under an optical microscope (Zeizz Axioskop, Oberkochen, Germany).
The cells (2×106) were resuspended in 1.5 mL lysis buffer [10 mM Tris (pH 7.5), 10 mM EDTA (pH 8.0), 10 mM NaCl, and 0.5% SDS] containing proteinase K (200 µg/mL). After overnight incubation at 48℃, 200 µL of ice-cold 5 M NaCl was added to the suspended cells and fragmented DNA was collected by ethanol precipitation. Approximately 7 µg of DNA from each sample was electrophoresed through a 2% agarose gel containing 0.5 µg/mL ethidium bromide. The DNA was visualized under UV light and photographed.
The cells were harvested with a cold cell lysis buffer (RIPA solution; 10 mM Tris, 150 mM NaCl, 1% Triton X-100, 1% deoxycholate, 0.1% SDS, 5 mM EDTA, pH 7.4) and the protein concentration was determined using the Brad-Ford protein assay. Cell lysates (50 µg) were used for SDS-PAGE coupled with western blot analysis. Antibodies against p53, Cdc2, cyclin B1 and beta-actin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA) and anti-mouse or anti-rabbit HRP- conjugated secondary IgG (Amersham Biosciences, Little Chalfont, UK) were used as a secondary antibody. Detection was carried out by incubating the membrane with an enhanced chemililuminescence reagent kit (Amersham Biosciences, Little Chalfont, UK).
Plasmids expressing GST-tagged azurin were cloned as indicated in the Materials and Methods section. As Fig. 1A. shows, this construct harbored the GST-tagged full-length azurin gene amplified from P. aeruginosa. This construct was transformed into an E. coli strain, BL21 (DE3), in order to obtain purified azurin. Fig. 1B. shows the final product of the purification procedure: 14 kDa azurin, which was pure enough to be used for cell treatment assays.
To elucidate the effects of purified azurin on OSCC, YD-9 cells were subjected to azurin treatment and their growth was monitored (Fig. 2A). Azurin decreased the viability of YD-9 cells in a dose-dependent manner, and 200 µg/mL azurin reduced cell growth by up to 50%. In the next step, the cell cytotoxic activity of purified azurin was tested at various time points. Interestingly, treatment of MG-63 osteosarcoma cells with 200 µg/mL azurin had no effect on growth, whereas treatment of YD-9 cells with 200 µg/mL azurin efficiently inhibited growth in a time-dependent manner. The decreased growth rates of YD-9 cells were about 25%, 50%, and 60% at 24, 48, and 72 h, respectively.
As Fig. 3A. shows, treatment with azurin effectively decreased the total number of cells and was accompanied by shrinkage and shape changes. When the cells were stained using the hemacolor staining method, which can discriminate between apoptosis and necrosis (Fig. 3B), condensed nuclei and an increase in apoptotic bodies were observed in azurin-treated cells. This phenotype was also observed when the nuclei of cells were stained with Hoechst 33342 (Fig. 3C). These results imply that azurin can induce apoptosis of YD-9 cells; therefore, DNA fragmentation was examined after azurin treatment. Fig. 3D shows that azurin treatment for 24 h was enough to induce DNA fragmentation.
In order to understand the molecular mechanisms inducing apoptosis after azurin treatment, the level of proteins involved in cell cycle regulation and apoptosis were monitored (Fig. 3E). The cellular p53 level increased within 8 hours of treatment and then decreased slightly in a time-dependent manner. Levels of Cdc2, which forms a complex with cyclin B1 during the G2/M transition, did not change with azurin treatment, but the level of cyclin B1 dramatically increased.
Since no anticancer drug developed to date can destroy all cancer cells, the effects of treatment with azurin in combination with other anticancer drugs were tested. 5-FU is one of the most frequently used drugs for treating oral cancer. Because of the variation in sensitivity of tumor cells to 5-FU, the effect of 5-FU on YD-9 and MG-63 cells was tested (Fig. 4A). Treatment of YD-9 cells and MG-63 cells with 5-FU alone resulted in approximately 30% growth inhibition at 1 mM. In contrast, when 5-FU was administered in combination with azurin, 10 µM 5-FU was enough to cause 80-90% growth inhibition. The synergistic effect of azurin on etoposide-induced apoptosis was also confirmed (Fig. 4B).
Here, we demonstrate the effects of azurin as an anti-oral cancer drug. The MTT assay showed that azurin effectively inhibits YD-9 oral cancer cell growth (Fig. 2A). Azurin treatment also increased the number of cells with apoptotic phenotypes (Fig. 3A-D). On the other hand, azurin treatment did not cause cytotoxicity of MG-63 osteosarcoma cells (Fig. 2B). This result was further confirmed by BrdU assay (data not shown). Another group reported that azurin had no effect on MG-63 cells, and they argued that this was caused by the lack of p53 in MG-63 cells.7 Fig. 3E shows that treating YD-9 cells with azurin enhances p53 expression, indicating that this increase in p53 might cause cell death. Interestingly, along with p53, expression of the cell-cycle regulator cyclin B1 was also enhanced by azurin treatment. The possible role of cyclin B1 in several DNA-damaging processes that induce apoptosis has been suggested. Cyclin B1 expression has been reported to be induced and this protein is reported to be active during stress-induced apoptosis in varieties of cancer cells, indicating that cyclin B1 participates in apoptosis.8-10 In keeping with these earlier findings, our data show that azurin effectively induces DNA fragmentation of YD-9 cells (Fig. 3D) along with increased expression of cyclin B1, indicating that increased cyclin B1 may play a role in azurin-induced apoptosis. The same set of experiments as in Fig. 3 was also performed using MG-63 cells after azurin treatment, but there were no significant differences (Data not shown).
In the final step of this study, we investigated azurin's ability to sensitize YD-9 and MG-63 cells to anticancer drugs. 5-FU was initially used in chemotherapy for gastrointestinal, breast, pancreas, and skin neoplasms. Although 5-FU remains a widely used chemotherapeutic agent for head and neck squamous cell carcinoma (HNSCC), its clinical importance has been decreasing in recent times because a number of HNSCCs are resistant to this agent.11 As shown in Fig. 4A, YD-9 and MG-63 cells have a strong resistance to 5-FU. When 5-FU was administered in combination with azurin, the viability of both cell lines was greatly decreased. Given that azurin alone does not have a cytotoxic effect on p53-null MG-63 cells, this result indicates azurin can enhance the sensitivity of MG-63 cells to 5-FU in a p53-independent manner. This sensitizing ability of azurin was further confirmed by combination treatment with etoposide (Fig. 4B). According to a report by Gomez, et al.,12 increased expression of cyclin B1 sensitizes cancer cells to chemotherapy-induced apoptosis. Therefore, the increase in cyclin B1 levels induced by azurin could be part of the molecular mechanism underlying azurin-induced sensitization of YD-9 and MG-63 cells to 5-FU and etoposide. Given the results of this study, future investigation is required.
Figures and Tables
Fig. 1
Extraction and purification of azurin from bacteria: (A) A schematic diagram of GST-tagged full length azurin (B) Expression of purified azurin protein: GST-azurin protein was amplified in bacteria and purified, as indicated in Materials and Methods. M: protein marker.

Fig. 2
Azurin inhibits the viability of YD-9 cells. (A) YD-9 cells were treated with various concentrations of azurin as indicated. After 48 h of incubation, cell growth was tested. Data shown are the average of 4 independent experiments, with a mean value of p<0.03. (B) YD-9 and MG-63 cells were treated with 200 µg/mL azurin and incubated for 24, 48, and 72 h as indicated, and cell viability was tested. Mean value, p<0.04.
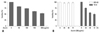
Fig. 3
Azurin triggers apoptosis of YD-9 cells. Morphological changes in YD-9 cells after 24 h incubation in the presence or absence of 200 µg/mL azurin, as observed with simple microscopy (A) and after cell staining with hemacolor (B) or Hoechst 33352 (C). (D) DNA fragmentation of 200 µg/mL azurin-treated cells occurred in a time-dependent manner. (E) YD-9 cells were exposed to 200 µg/mL azurin for indicated times. Total protein (30 µg) was subjected to SDS-PAGE and western blotting. Data shown is the representative of 3 independent experiments. M, DNA marker; C, Control (lysate of untreated cells).
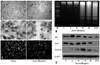
Fig. 4
Azurin enhances sensitivity of YD-9 and MG-63 cells to 5-fluorouracil and etoposide. YD-9 and MG-63 cells were treated with 10-1,000 µM of 5-fluorouracil (5-FU) (A) or 2-10 µg/mL etoposide (B) along or not along with 200 µg/mL azurin as indicated for 24 h and subjected to a cell growth assay. Data shown are representative of 4 independent experiments, with p<0.02.

ACKNOWLEDGEMENTS
This study was financially supported by Medical Research Institute Grant (2008-0025000), Pusan National University.
References
1. Hooper SJ, Wilson MJ, Crean SJ. Exploring the link between microorganisms and oral cancer: a systematic review of the literature. Head Neck. 2009. 31:1228–1239.

2. Chen XX, Lai MD, Zhang YL, Huang Q. Less cytotoxicity to combination therapy of 5-fluorouracil and cisplatin than 5-fluorouracil alone in human colon cancer cell lines. World J Gastroenterol. 2002. 8:841–846.

3. Yamada T, Fialho AM, Punj V, Bratescu L, Gupta TK, Chakrabarty AM. Internalization of bacterial redox protein azurin in mammalian cells: entry domain and specificity. Cell Microbiol. 2005. 7:1418–1431.

4. Yamada T, Goto M, Punj V, Zaborina O, Chen ML, Kimbara K, et al. Bacterial redox protein azurin, tumor suppressor protein p53, and regression of cancer. Proc Natl Acad Sci U S A. 2002. 99:14098–14103.

5. Punj V, Bhattacharyya S, Saint-Dic D, Vasu C, Cunningham EA, Graves J, et al. Bacterial cupredoxin azurin as an inducer of apoptosis and regression in human breast cancer. Oncogene. 2004. 23:2367–2378.

6. Yamada T, Goto M, Punj V, Zaborina O, Kimbara K, Das Gupta TK, et al. The bacterial redox protein azurin induces apoptosis in J774 macrophages through complex formation and stabilization of the tumor suppressor protein p53. Infect Immun. 2002. 70:7054–7062.

7. Yang DS, Miao XD, Ye ZM, Feng J, Xu RZ, Huang X, et al. Bacterial redox protein azurin induce apoptosis in human osteosarcoma U2OS cells. Pharmacol Res. 2005. 52:413–421.

8. Borgne A, Versteege I, Mahé M, Studeny A, Léonce S, Naime I, et al. Analysis of cyclin B1 and CDK activity during apoptosis induced by camptothecin treatment. Oncogene. 2006. 25:7361–7372.

9. Porter LA, Singh G, Lee JM. Abundance of cyclin B1 regulates gamma-radiation-induced apoptosis. Blood. 2000. 95:2645–2650.
10. Porter LA, Cukier IH, Lee JM. Nuclear localization of cyclin B1 regulates DNA damage-induced apoptosis. Blood. 2003. 101:1928–1933.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download