Abstract
During the 2009 novel influenza (H1N1) pandemic, the sensitivity of direct immunofluorescence assay (DFA) for H1N1 infection was 62% (266/429) of that of the polymerase chain reaction (PCR) test. The sensitivity of the DFA differed significantly with the age of patients: the sensitivity was the highest (71.8%) for patients aged <10 years and the lowest for patients aged ≥30 years. The sensitivity of DFA in patients aged ≥30 years was 40.7%. Furthermore, the sensitivity (67.3%, 171/254) of DFA was higher for patients who had a high temperature at admission. An increase in the incidence of H1N1 infection did not influence the sensitivity of DFA (62.1% vs. 62%; p=0.984) test, but resulted in a decrease in the negative predictive value, from 92.4% (700/757) to 69.6% (247/355). PCR may be useful as the initial test for diagnosing H1N1 infection in patients aged ≥30 years with a normal temperature at presentation.
In the light of global outbreak of the novel influenza A (H1N1) virus infection, rapid diagnosis is important for the timely initiation of antiviral therapy and implementation of infection control strategies. Since the signs and symptoms of influenza are similar to other respiratory viral infections, the differential diagnosis of influenza may be difficult when based solely on clinical symptoms. Rapid influenza antigen tests may prove useful because of their quick results and technical simplicity. However, these tests have low to moderate sensitivity.1 The direct immunofluorescence assay (DFA) is a rapid test, which yields results within 4 h. The sensitivity of DFA for seasonal influenza is higher than that of the rapid antigen test.2,3 Polymerase chain reaction (PCR) is considered as the confirmatory test for the diagnosis of the novel influenza A (H1N1) virus infection because of its high sensitivity and specificity. In Korea, during the early stage of the pandemic, PCR for H1N1 virus was available only in designated laboratories. With the spread of the H1N1 epidemic to all parts of the nation, the government added more centers to conduct the PCR test for H1N1 infection and had temporarily provided medical insurance to patients for PCR tests.
Our hospital (Wonkwang University Hospital) was designated as one of these centers on August 17, 2009. In our hospital, PCR for H1N1 and the DFA test were performed 3 times a day. We evaluated the diagnostic accuracy of DFA in the detection of H1N1 infection and potential factors that might have influenced the results of DFA.
From August 2009 to October 2009, a total of 2,310 patients were tested for H1N1 infection; 2,302 by reverse transcription PCR (RT-PCR) and 1,392 by DFA. Among the 2,310 patients tested, 1,383 patients underwent PCR and DFA tests concurrently for the detection of the H1N1 virus. The study subjects were classified into four age categories: below 10 years, 10-19 years, 20-29 years, and ≥30 years. All specimens were collected from the posterior nasopharynx by using flocked swabs (Copan Diagnostics) and transported to the microbiology laboratory in viral transport medium (Remel). Specimens were stored at 4℃ until further processing. D3 Respiratory Virus Reagents (Diagnostic Hybrids, Athens, OH, USA) were used for the DFA. A positive result was defined as the detection of 2 or more intact cells exhibiting a specific fluorescence pattern. PCR testing for H1N1 infection was performed using the New InfA (H1N1) & InfA real-time RT-PCR kit (BIONEER CO., Daejeon, Korea), in accordance with the protocol by the Center for Disease Control and Prevention, Influenza Branch. The specimens for DFA and RT-PCR were assessed separately. We calculated the sensitivity, specificity, positive predictive value and negative predictive values for the DFA test by using standard formulae. The SPSS software (version 15.0) was used for statistical analysis and a p-value of less than 0.05 was considered statistically significant.
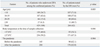
During the study period, H1N1 infection was detected in 907 patients (39.4%, 907/2302) by the PCR test. In this group of patients, 436 were tested simultaneously by DFA. However, 7 patients whose medical records could not be reviewed were excluded from the study. Finally, 429 patients amongst the 907 were included in this study. In the group of 429 patients, 63.4% were males, and the mean age of the subjects was 14.6±10.1 years. The highest proportion of subjects were 10-19 years, followed by those aged <10 years, then those aged 20-29 years, and finally those aged ≥30 years. The sensitivity of DFA assessed in comparison with the PCR test results was 62% (266/429). The sensitivity of DFA differed significantly with patient's age: 71.8% (107/149) in subjects aged <10 years, 57.8% (108/187) in patients aged 10-19 years, 60.6% (40/66) in patients aged 20-29 years, and 40.7% (11/27) in patients aged ≥30 years. Using multiple logistic regression, the sensitivity of DFA was found to be correlated significantly with the patient's temperature at admission (odds ratios 1.241, 95% confidence interval 1.005-1.532; p=0.045). The number of H1N1 virus infection by PCR test increased as follows: 13 cases in August, 77 cases in September, 58 cases in the first half of October and 759 cases in the second half of October. However, the sensitivity (62.1% vs. 62%; p=0.984) of DFA was not influenced by the increase of the incidence (Table 1).
In our study, assuming RT-PCR as the "gold standard", the sensitivity of the DFA test was 62% (266/429), the specificity was 100% (947/947), the positive predictive value was 100% (266/266), and the negative predictive value was 85.2% (947/1112). The sensitivity (71.8%, 107/149) of DFA was the highest for patients aged <10 years, and the lowest (40.7%, 11/27) for patients aged ≥30 years. Compared with a previous study,4 which primarily included adult subjects (mean age, 44 years), our study showed a noticeably lower sensitivity (40.7%, 11/27) for adult patients. The use of flocked swabs enabled the collection of adequate specimens, as reported previously.5 Among the specimens collected in this study, only two (2/1392) had inadequate number of cells for DFA test. Furthermore, a serial process, of which the specimens were collected from patients with ILI, then transported to the microbiology laboratory, and stored at 4℃, was completed within a mean time of 40 minutes. Previous study reported that symptom severity score at presentation correlated positively with the viral load and duration of viral shedding in adult patients with influenza.6 In our study, the body temperature at admission correlated negatively with the cycle threshold (Ct) values of the RT-PCR in adult patients (p<0.001). The lower the Ct value, the positivity of DFA test increased significantly (p<0.001) (Table 2). The sensitivity of DFA for patients with ≥37.8℃ was higher (67.3%, 171/254) than in patients (54.3%, 95/175) with <37.8℃ at admission. The mean body temperature of the subjects aged ≥30 years was significantly lower (37.2±0.65℃) than that of patients in other age groups (38.2±0.95℃, 37.9±0.99℃ and 37.8± 0.65℃ for age group <10 years, 10-19 years, 20-29 years, respectively; p<0.001). These patients might have taken medicines before visiting the hospital, which might have influenced the results of DFA. In addition, we examined the test performance not only for patients with influenza-like illnesses (18/27) but also for those who had few symptoms (9/27) but wanted to be tested for H1N1 infection. During our study period, a pandemic of H1N1 infection was declared in week 43 (October 18). The sensitivity (62%, 176/284) of the DFA test was not influenced by an increase in the incidence of the infection, but the negative predictive value of the test was reduced. The negative predictive value is specific to the disease prevalence associated with the period of epidemic.4 In our study, the overall negative predictive value was 85.6% (947/1112), but decreased to 69.6% (247/355) after the onset of the pandemic.
The mean turn around time of DFA and RT-PCR was 8 h 33 min and 9 h 37 min, respectively.
The minimal difference of turn around time between the DFA test and RT-PCR, the provision of medical insurance cover for the PCR test and the limited role of the DFA in the exclusion of H1N1 prompted us to discontinue the use of the DFA test after October 31, 2009.
Figures and Tables
Notes
References
1. Vasoo S, Stevens J, Singh K. Rapid antigen tests for diagnosis of pandemic (Swine) influenza A/H1N1. Clin Infect Dis. 2009. 49:1090–1093.

2. Steininger C, Redlberger M, Graninger W, Kundi M, Popow-Kraupp T. Near-patient assays for diagnosis of influenza virus infection in adult patients. Clin Microbiol Infect. 2009. 15:267–273.

3. Habib-Bein NF, Beckwith WH 3rd, Mayo D, Landry ML. Comparison of SmartCycler real-time reverse transcription-PCR assay in a public health laboratory with direct immunofluorescence and cell culture assays in a medical center for detection of influenza A virus. J Clin Microbiol. 2003. 41:3597–3601.

4. Pollock NR, Duong S, Cheng A, Han LL, Smole S, Kirby JE. Ruling out novel H1N1 influenza virus infection with direct fluorescent antigen testing. Clin Infect Dis. 2009. 49:e66–e68.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download