Abstract
Purpose
PCR is widely used for rapidly and accurately detecting Mycobacterium Species. The purpose of this study was to assess the diagnostic performance of three real-time PCR kits and evaluate the concordance with two older PCR methods.
Materials and Methods
Using 128 samples, the five PCR methods were assessed, including an in-house PCR protocol, the COBAS Amplicor MTB, the COBAS TaqMan MTB, the AdvanSure TB/NTM real-time PCR, and the Real-Q M. tuberculosis kit. The discrepant results were further examined by DNA sequencing and using the AdvanSure Mycobacteria Genotyping Chip for complete analysis.
Results
For Mycobacterium tuberculosis (MTB) detection, all five kits showed 100% matching results (positive; N = 11 and negative; N = 80). In non-tuberculous mycobacterium (NTM) discrimination, the AdvanSure yielded two true-positive outcomes from M. intracellulare and one false positive outcome, while the Real-Q resulted in one true-positive outcome and one false negative outcome for each case and another false negative result using the provided DNA samples.
Conclusion
Real-time PCR, yielded results that were comparable to those of the older PCR methods for detecting MTB. However, there were disagreements among the applied kits in regard to the sample test results for detecting NTM. Therefore, we recommend that additional confirmatory measures such as DNA sequencing should be implemented in such cases, and further research with using a larger numbers of samples is warranted to improve the detection of NTM.
Tuberculosis (TB) is a major global public health problem and one of the leading infectious causes of death. According to a recent WHO report, 1.6 million people worldwide died of TB in 2005.1 Korea has shown a higher prevalence of mycobacterial infection than most of the other developed countries, and the prevalence of non-tuberculous mycobacterium (NTM) infection has increased in the developed countries.2 Therefore, an increased emphasis has currently been placed on the rapid identification of Mycobacterium tuberculosis (MTB) and NTM.2,3
The introduction of the nucleic acid amplification techniques has been one of the major developments in diagnosis of mycobacterium, and real-time PCR has recently been proposed for the detection.4 Real-time PCR has several advantages, such as a short turnaround time, a low contamination rate due to the use of a closed system and the ability to quantify the bacterial load.4,5 In Korea, two real-time PCR kits that simultaneously detect MTB and NTM have been developed for rapid MTB detection and NTM discrimination. However, it is unclear whether these real-time PCR kits offer the same sensitivity and specificity as conventional PCR. In this study, we evaluated the diagnostic performance of three real-time PCR kits, by comparing the results from these kits with those from our in-house PCR and the conventional qualitative PCR techniques. The discrepant results were confirmed by DNA sequencing and genotyping chip analyses.
This study was conducted at Kyung Hee Medical Center, Seoul, Korea from September 2009 to October 2009, using 91 clinical specimens and 37 provided DNA samples (Table 1). The clinical specimens contained pulmonary and extrapulmonary specimens, and the smear positive and negative specimens (Table 2). The specimens were stored at - 70℃ in the state of extracted DNA.
After mixing 3 mL of the specimen sample with an equal amount of 1N NaOH, the samples were vortexed for 30 seconds and then left at room temperature for 15 minutes. Subsequently, the mixture was centrifuged for 20 minutes at 3,000 rpm. After discarding the supernatant, the pellet was mixed with 1 mL of a buffer, vortexed and centrifuged for 5 minutes at 7,000 rpm. This process was repeated three times. Once the supernatant was completely removed, the pellet was dissolved in an adequate amount (50-200 µL) of 5% Chelex100 with a buffer. The sample was then boiled for 10 minutes and centrifuged for 5 minutes at 12,000 rpm. In the final supernatant, 1-2 µL was taken for the PCR reaction.
One hundred µL of the prepared sample was added to 500 µL of Respiratory Specimen Wash Solution, and this was centrifuged for 10 minutes at 12,500 rpm. After removing the supernatant, 100 µL of Respiratory Specimen Lysis Solution was added and this was vortexed until the pellet was dissolved. The sample was incubated for 45 minutes at 60℃, and then 100 µL of Respiratory Specimen Neutralization Solution was added. Using 50 µL of this reaction mixture with a 50 µL master mix, a PCR reaction was conducted in a reaction tube.
The primer sequences used for in-house PCR have previously been described.4 The in-house PCR products were determined to be positive if there was a 285 bp-sized band on the gel electrophoresis (Fig. 1). Using the IS6110 primer, nested PCR was conducted using the GeneAmp PCR system 9600 (Perkin Elmer, CT, USA). For the COBAS Amplicor, which is based on the PCR-hybridization method, Amplicor (Roche Molecular Systems) was used and the sample was determined to be positive if absorbance was equal to or greater than 0.35. In addition, the COBAS TaqMan MTB (Roche, Branchburg, NJ, USA) was used with the COBAS TaqMan 48 Analyzer (Roche), the AdvanSure TB/NTM real-time PCR kit (LG Lifescience, Seoul, Korea) with the SLAN real-time PCR detection system (LG Lifescience), and the Real-Q M. tuberculosis kit (Biosewoom, Seoul, Korea) with the Rotor-Gene Q (Corbett Life Science, Sydney, Australia), for the real-time PCR analysis.
To solve inconsistent results, the 16S-23S internal transcribed spacer (ITS) was sequenced and the results were confirmed by a BLAST search. In addition, the AdvanSure Mycobacteria Genotyping Chip (LG Lifescience, Seoul, Korea) was used in order to reconfirm the results. The AdvanSure Mycobacteria Genotyping Chip implements the hybridization method by employing the ITS probe that exists between the 16S rRNA and 23S rRNA of mycobacteria, thereby identifying both MTB and NTM. The chip was used according to the manufacturer's instructions; hybridization was conducted after PCR and the results were analyzed with the provided insert.
Out of the 91 clinical samples, there were 75 pulmonary samples (64 sputum samples, 11 BAL samples) and 16 non-pulmonary samples. The results from the two previously implemented PCR methods showed a perfect match for all 91 samples, including the 11 MTB positive samples. These were found to be positive by the other three real-time PCR techniques, and 100% concordance was observed among all the tests (Table 3). One of the pulmonary specimens that 5 PCR methods detected MTB showed negative in the TB culture and the acid fast bacilli (AFB) stain. He presented blood-tinged sputum and multiple haziness in the chest X-ray film. Based on clinical manifestation and the PCR result, TB was diagnosed.
By examining 80 clinical samples that were found to be negative for NTM on at least one test and 37 DNA samples that were provided, we compared the results of the AdvanSure and the Real-Q, which detected NTM. Out of 80 MTB negative samples, one sample was identified as positive by both kits. It was a sputum sample that was later confirmed to contain M. intracellulare by subsequent DNA sequencing. For 2 other samples of the 80 samples, the two kits yielded opposite results; the result from AdvenSure was positive while that from Real-Q was negative in both cases (Table 4). In one of the pulmonary specimens with NTM negative results by 2 PCR methods showed positive in the culture and the AFB stain. It was then identified to be M. abscessus by PCR-restriction fragment length polymorphism (RFLP) in the reference laboratory.6
These clinical samples were further analyzed through DNA sequencing, genotyping chip tests, AFB stains, and cultures. Interestingly, it was concluded that one sample was NTM false positive by AdvanSure, while the other sample was NTM false negative by the Real-Q (M. intracellulare)(Table 4). Therefore, two samples out of 80 clinical samples that were MTB negative were found to be NTM positive, representing 2.5% of the total samples (Fig. 2). In the chart review, these two patients were confirmed to have NTM TB as well.
The 37 DNA samples provided by LG Lifescience were used to evaluate these two kits by blind testing. Because both kits showed negative for all 6 non-NTM DNA samples, there were no false positive results. Nevertheless, there was one false negative by the Real-Q, which was negative for M. lentiflavum.
Considering the fact that many laboratories have recently implemented real-time PCR assays to detect viruses or fastidious microbes, and that real-time PCR uses a closed system, thereby lowering the potential for false positive results caused by cross-contamination of the amplified products, it is highly expected that the usage of real-time PCR will continue to increase in the field of microbiological diagnosis.7-9 In addition, the need to differentiate MTB and NTM is gaining more importance, owing to the growing occurrence of NTMs and the fact that both MTB and NTM require different medications.2 In the present study, therefore, we compared and assessed the diagnostic performance of three types of commercial real-time PCR kits for MTB detection and NTM differentiation, as well as two PCR methods previously implemented in our hospital.
Since the COBAS Amplicor test was shown to have a low sensitivity for non-respiratory samples, Kyung Hee University Medical Center had been using the Amplicor for respiratory samples while in-house PCR was used for non-respiratory samples.10 In our previous reports, the diagnostic sensitivity and specificity of our in-house PCR method were shown to be 81.0% and 99.6%, respectively.11 For the COBAS Amplicor, the diagnostic sensitivity varies from about 70% to about 80%-90%, depending on the studies.11,12 One study in Korea showed the sensitivity and the specificity of the AdvanSure to be 97.9% and 100%, respectively, whereas 91% and 87%, respectively, in the other study, although there are only a fewer reports on the sensitivity and specificity of TB detection by real-time PCR.12,13 These two studies are different in their sample composition; the former used only respiratory samples that comprised sputum and bronchoalveolar lavage (BAL), while the latter included non-respiratory samples such as body fluid and urine, which made up 30.5% of the sample group.13,14 Therefore, it is highly possible that one of the causes for the different estimates of sensitivity and specificity is the difference in the sample composition. In particular, PCR inhibitory materials in non-respiratory samples can result in a lower sensitivity when performing real-time PCR on those samples.15,16
We found a 100% match of the results for MTB detection in a comparative analysis of five PCR methods, as well as several discordant results with culture and AFB stain. In one of the sputum specimens which showed MTB positive in five PCR methods, MTB did not grow and AFB stain revealed no acid fast bacili. Because the patient had the medical history of TB and showed clinical symptoms and signs to be compatible with active TB, the medication for MTB was started based on the PCR result.
There were also a few discrepant results regarding the detection of NTM. After detecting NTM in the MTB PCR-negative clinical samples with the AdvanSure and the Real-Q, two cases showed mismatching results, while one other case turned out to be positive with both kits. In the two mismatching cases, the results from the AdvanSure were all positive and the results from the Real-Q were all negative. The medical records indicated that one of 2 AdvanSure positive cases showed a history of NTM TB treatment, and the offending organism was later identified to be M. intracellulare by DNA sequencing and genotyping chip tests. The other AdvanSure positive case turned out to be TB false positive after additional analysis, thus concluding that there was one false positive result by the AdvanSure and one false negative result by the Real-Q. The false positive sample in the AdvanSure was a sputum sample from a 13-year-old male patient who had a history of TB before the age of 1 and he had undergone a 6-month medication. That patient was hospitalized for symptoms of pneumonia, and his sputum test was negative after an AFB stain. Since the patient did not have clinical evidence of TB, he was released after being treated for bacterial pneumonia. The Ct value for this sample was 33.25, which was higher than 27.02 of the internal control. The sample that turned out to be false negative in the Real-Q was a sputum sample that continuously indicated a positive value of 2+ to 3+ in AFB stain and the offending organism had been identified as M. intracellulare by culture. Both samples came from patients who were diagnosed with NTM-caused TB, with both being identified as M. intracellulare and the 2 samples were found negative by the TaqMan. Among NTM PCR-negative samples in the AdvanSure and the Real-Q, there was a sputum specimen that showed positive in the culture and the AFB stain. We referred the NTM identification to the reference laboratory, and it was identified to be M. abscessus.6 In conclusion, three NTM true-positive results out of 80 MTB negative samples (2.5%) were confirmed. As for 37 DNA samples, the AdvanSure and the Real-Q recognized 6 non-NTM DNA (MTB, nocardial and rhodococcal DNA) as negative, while the Real-Q recognized M. lentiflavum as negative out of 31 NTM DNA samples. Considering these results, the implementation of real-time PCR methods that are capable of NTM differentiation made it possible to make a swift and accurate diagnosis of NTM TB.
In conclusion, the five kits included in this comparative analysis yielded good matching ratios for the MTB detection results. However, for detecting NTM, there was some discordance, therefore, further confirmatory measures such as DNA sequencing, a medical record review, and/or culture are required in order to make an accurate diagnosis. Future research on this topic is certainly needed, and this research would be benefited from the putatively increased NTM detection capacities to assess numerous and varied samples, including non-respiratory samples or negative samples in ATB stain.
Figures and Tables
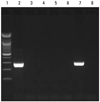 | Fig. 1In-house MTB PCR. Lane 1, size marker; Lane 2, internal control; Lane 7, positive; Lane 3-6, 8, negative. MTB, Mycobacterium tuberculosis. |
 | Fig. 2Two discrepant cases reaffirmed by sequencing and using the AdvanSure Mycobacteria Genotyping Chip (LG Life science, Seoul, Korea). (A) For 37 DNA samples provided, the AdvanSure kit detected all mycobacterial species and Real-Q kit showed a negative result for M. lentiflavum (target sequence; IS6110 region). (B) In contrast with the one false positive result from the AdvanSure kit, the Real-Q missed one true positive M. intracellulare in the clinical samples (target sequence; 16S-23S internal transcribed spacer region). |
Table 3
The MTB PCR Results for 91 Clinical Specimens

MTB, Mycobacterium tuberculosis; AFB, acid fast bacilli.
*COBAS Amplicor MTB (Roche, Indianapolis, IN, USA).
†COBAS TaqMan MTB (Roche, USA).
‡AdvanSure TB/NTM real-time PCR kit (LG Lifescience, Seoul, Korea).
§Real-Q M. tuberculosis kit (Biosewoom, Seoul, Korea).
∥One of the 91 clinical samples had no culture and AFB stain results.
ACKNOWLEDGEMENTS
We thank Prof. Tae Sung Park, at our department for his invaluable supports to this manuscript.
References
1. World Health Organization. New technologies for tuberculosis control: a framework for their adoption, introduction and implementation. World Health Organization, 2007. Appia, Geneva, Switzerland: WHO Press.
2. Chang HE, Heo SR, Yoo KC, Song SH, Kim SH, Kim HB, et al. [Detection of Mycobacterium tuberculosis complex using real-time polymerase chain reaction]. Korean J Lab Med. 2008. 28:103–108.

3. Kang SH, Yoo KC, Park KU, Song J, Kim EC. [Usefulness of multiplex real-Time PCR and melting curve analysis in identification of nontuberculous mycobacteria]. Korean J Lab Med. 2007. 27:40–45.

4. Baba K, Pathak S, Sviland L, Langeland N, Hoosen AA, Asjo B, et al. Real-time quantitative PCR in the diagnosis of tuberculosis in formalin-fixed paraffin-embedded pleural tissue in patients from a high HIV endemic area. Diagn Mol Pathol. 2008. 17:112–117.

5. Lee MF, Chen YH, Peng CF. Evaluation of reverse transcription loop-mediated isothermal amplification in conjunction with ELISA-hybridization assay for molecular detection of Mycobacterium tuberculosis. J Microbiol Methods. 2009. 76:174–180.

6. Lee H, Park HJ, Cho SN, Bai GH, Kim SJ. Species identification of mycobacteria by PCR-restriction fragment length polymorphism of the rpoB gene. J Clin Microbiol. 2000. 38:2966–2971.

7. Flores E, Rodríguez JC, Garcia-Pachón E, Soto JL, Ruiz M, Escribano I, et al. Real-time PCR with internal amplification control for detecting tuberculosis: method design and validation. APMIS. 2009. 117:592–597.

8. Richardson ET, Samson D, Banaei N. Rapid Identification of Mycobacterium tuberculosis and nontuberculous mycobacteria by multiplex, real-time PCR. J Clin Microbiol. 2009. 47:1497–1502.

9. Mackay IM. Real-time PCR in the microbiology laboratory. Clin Microbiol Infect. 2004. 10:190–212.

10. Tortoli E, Tronci M, Tosi CP, Galli C, Lavinia F, Natili S, et al. Multicenter evaluation of two commercial amplification kits (Amplicor, Roche and LCx, Abbott) for direct detection of Mycobacterium tuberculosis in pulmonary and extrapulmonary specimens. Diagn Microbiol Infect Dis. 1999. 33:173–179.

11. Yang HY, Lee HJ, Park SY, Lee KK, Suh JT. [Comparison of In-house Polymerase Chain Reaction Assay with Conventional Techniques for the Detection of Mycobacterium tuberculosis]. Korean J Lab Med. 2006. 26:174–178.

12. Choi YM, Lee MH. Detection of Mycobacterium tuberculosis in sputum by using polymerase chain reaction. Korean J Clin Microbiol. 1999. 2:144–151.
13. Kim YJ, Park MY, Kim SY, Cho SA, Hwang SH, Kim HH, et al. [Evaluation of the performances of advanSure TB/NTM real time PCR Kit for detection of mycobacteria in respiratory specimens]. Korean J Lab Med. 2008. 28:34–38.

14. Jung CL, Kim MK, Seo DC, Lee MA. Clinical usefulness of real-time PCR and amplicor MTB PCR assays for diagnosis of tuberculosis. Korean J Clin Microbiol. 2008. 11:29–33.

15. Scarparo C, Piccoli P, Rigon A, Ruggiero G, Scagnelli M, Piersimoni C. Comparison of enhanced Mycobacterium tuberculosis amplified direct test with COBAS AMPLICOR Mycobacterium tuberculosis assay for direct detection of Mycobacterium tuberculosis complex in respiratory and extrapulmonary specimens. J Clin Microbiol. 2000. 38:1559–1562.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download