Abstract
Purpose
Antimicrobial resistance monitoring could be a useful source of information for treating and controlling nosocomial infections. We analyzed antimicrobial resistance data generated by Korean Hospitals and by a commercial laboratory in 2005 and 2007.
Materials and Methods
Susceptibility data for 2005 and 2007 were collected from 37 and 41 hospitals, respectively, and from one commercial laboratory. Intermediate susceptibility was not included in the calculation of resistance rates.
Results
Methicillin-resistant Staphylococcus aureus (MRSA) (64%), third-generation cephalosporin-resistant Klebsiella pneumoniae (29%), fluoroquinolone-resistant Escherichia coli (27%), Pseudomonas aeruginosa (33%), and Acinetobacter spp. (48%), and amikacin-resistant P. aeruginosa (19%) and Acinetobacter spp. (37%) were prevalent in hospitals in 2007. A gradual increase of vancomycin-resistant Enterococcus faecium and imipenem-resistant Acinetobacter spp. was observed. Higher incidences of third-generation cephalosporin-resistant E. coli and K. pneumoniae and imipenem-resistant P. aeruginosa were found in the commercial laboratory than in the hospitals.
Conclusion
Methicillin-resistant S. aureus, third-generation cephalosporin-resistant K. pneumoniae, and fluoroquinolone-resistant E. coli, P. aeruginosa and Acinetobacter spp. remain prevalent in Korea, while the incidence of vancomycin-resistant E. faecium and imipenem-resistant Acinetobacter spp. has increased gradually. The higher prevalences of third-generation cephalosporin-resistant E. coli and K. pneumoniae, and imipenem-resistant P. aeruginosa in the commercial laboratory are a new concern.
An alarming increase in bacteria with resistance to various antibiotics has been reported in many parts of the world. However, the prevalence of antibiotic-resistant bacteria varies greatly from country to country, because it is influenced by antimicrobial usage and the spread of antibiotic resistance. With the increased incidence of antibiotic-resistant bacteria, optimal treatment of infected patients is often possible only by detection of the etiologic organism and determination of its susceptibility to various antibiotics. However, isolation of the infecting organism is not always feasible, and even if it is possible, results are not promptly accessible. Consequently, initial antibiotic therapy is generally empirical. The appropriate antibiotics can be empirically selected only with the full knowledge of the current regional resistance pattern.1 Antimicrobial resistance surveillance serves as a fundamental basis for appreciating trends in resistance, developing accurate treatment guidelines, and evaluating the efficacy of interventions (www.who.int/drugresistance/en/). Monitoring the temporal patterns of resistance is an essential element in the detection of subtle variations in antimicrobial resistance.2
Trends in antimicrobial resistance are most accurately predicted by collecting clinical isolates and testing their susceptibilities at a reference laboratory. This approach has some drawbacks, however: the number of characterized isolates is limited, and is costly. One example of this approach is the SENTRY program.1 Another frequently used surveillance method is the analysis of routine susceptibility test data from clinical laboratories. This approach has inherent inaccuracies, because not all laboratories use the same methods, but it has the advantage of not requiring extensive resources.3,4 Based on a WHO recommendation, the KONSAR program was initiated in 1997.5 Two surveillance methods are employed in the KONSAR program: annual analysis of test data generated by the participant laboratories6 and the testing of problematic organisms collected by the coordinating laboratory.7
Some of the most disturbing resistance trends found in the first program were high incidences of methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecium, and increases in the prevalence of third-generation cephalosporin-resistant Klebsiella pneumoniae and imipenem-resistant Pseudomonas aeruginosa and Acinetobacter spp. Some of these particular antibiotic-resistant organisms were also found to be prevalent by a commercial laboratory (ComLab).6 The second program revealed wide dissemination of metallo-β-lactamase (MBL)-producing Pseudomonas spp. and Acinetobacter spp.,8,9 and a rapid increase in OXA carbapenemase-producing Acinetobacter baumannii,9 as well as plasmid-mediated AmpC enzyme-producing Escherichia coli and K. pneumoniae.10 For this study, we compared the rank order of organisms and resistance rates in hospitals and in a ComLab.
Susceptibility data for 2005 and 2007 were collected from 37 and 41 hospitals, respectively, and from one ComLab. The KONSAR program participating hospitals have their own laboratories, whereas the ComLab in Seoul mostly tests specimens from smaller hospitals or clinics without in-house microbiology service. Of these, data from 18 and 23 hospitals, respectively, were excluded from the analysis because the data contained multiple isolates from a single patient.
In previous surveillance studies, dividing the hospitals into three groups according to location and bed capacity did not result in a significant difference in resistance rates. Therefore, all hospitals were grouped together in the present analysis. The test data obtained at the ComLab were analyzed separately. Intermediate susceptibility was not included in the calculation of resistance rates as was in the previous studies. For Streptococcus pneumoniae, the penicillin G breakpoint of meningitis was used, although most of the isolates were from other sites. If less than ten isolates of an organism in a hospital were reported, the organism was not included in the calculation of mean rates to avoid biasing these rates.11,12 Statistical significance of resistance rates was not analyzed in this surveillance, as it was considered difficult,13 and as it was the common practice in the large scale and continuous surveillance program.14,15
In 2007, E. coli ranked No. 1 in terms of the most prevalent organism isolated in both hospitals and the ComLab (19.3% and 27.1%) (Table 1). The second most prevalent organism was S. aureus in hospitals (17.8%) and P. aeruginosa in the ComLab (17.5%). The organisms ranked No. 3 through No. 5 in hospitals were coagulase-negative staphylococci (CNS) (13.1%), P. aeruginosa (11.7%), and K. pneumoniae (10.0%), whereas in the ComLab, these organisms were S. aureus (11.9%), K. pneumoniae (11.9%), and E. faecalis (8.4%). Changes in the rank of prevalent organisms in hospitals since 1998 were as follows: E. faecium from No. 10 (2.4%) to No. 7 (6.2%), and Acinetobacter spp. from No. 5 (9.5%) to No. 8 (5.3%).
The appropriateness of antimicrobial agents used to test susceptibility was analyzed for E. coli and S. aureus as representatives of Gram-negative bacilli and Gram-positive cocci, respectively. Overall, more hospitals followed the CLSI guidelines in 2007 than in 2002. However, the proportions of hospitals that used cephalothin, cefoxitin, ampicillin-sulbactam or amoxicillin-clavulanic acid, and cotrimoxazole to test the susceptibility of E. coli were only 86%, 81%, 65%, and 51%, respectively (data not shown). All hospitals used penicillin G, oxacillin (or cefoxitin to screen MRSA), erythromycin, and clindamycin to test the susceptibilities of S. aureus, but only 61% of hospitals used cotrimoxazole.
Surveillance results for both 2005 and 2007 (Tables 2-5) were analyzed in this study, but for simplicity, only the results for 2007 are described. Of the S. aureus hospital isolates, 64% and 8% were resistant to oxacillin (or to cefoxitin for MRSA detection) and to cotrimoxazole, respectively (Table 2). In the ComLab, 63% and 56% of S. aureus isolates were resistant to oxacillin and clindamycin, respectively. The resistance rates of CNS in hospitals to oxacillin and clindamycin were 64% and 29%, respectively, whereas the respective rates in the ComLab were 70% and 30%.
In the hospitals, when the meningitis breakpoint was applied, 69% of S. pneumoniae were nonsusceptible to oxacillin, suggesting penicillin nonsusceptibility (Table 3). The resistance of E. faecalis isolates to ampicillin and vancomycin were very low, but those of E. faecium were 86% and 21%, respectively. In the ComLab, 42% of S. pneumoniae were nonsusceptible to oxacillin, 86% of E. faecium were resistant to ampicillin, and 17% of E. faecium were resistant to vancomycin.
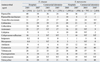
The resistance rates of E. coli in the hospitals to ampicillin, cefotaxime, amikacin, and fluoroquinolone were 60%, 11%, 2%, and 27%, respectively (Table 4). In the ComLab, the resistance rates of E. coli to ampicillin and amikacin were similar to those in the hospitals, but the rates of resistance to cefotaxime and to fluoroquinolone were higher: 45% and 42%, respectively. Of K. pneumoniae hospital isolates, 29% were resistant to ceftazidime, 21% to cefoxitin, 18% to amikacin, and 27% to fluoroquinolone (Table 4). In the ComLab, the resistance rates to ceftazidime, cefoxitin, amikacin, and fluoroquinolone were higher than those in the hospitals. ESBL-producing E. coli and K. pneumoniae were tested by 21 and 20 hospitals, respectively. The mean proportion of ESBL-producing E. coli and K. pneumoniae isolates was 12% and 29% in 2007, respectively (data not shown).
Of the Enterobacter cloacae hospital isolates, 30% were resistant to ceftazidime, 8% to cefepime, 6% to amikacin, and 10% to fluoroquinolone (Table 5). The resistance rates of this organism in the ComLab to ceftazidime and cefepime were slightly lower, whereas the rates of resistance to amikacin and fluoroquinolone were similar between the ComLab and hospitals. Of the Serratia marcescens isolates, 17% were ceftazidime-resistant, 10% were cefepime-resistant, 12% were amikacin-resistant, and 10% were fluoroquinolone-resistant (Table 5). In the ComLab, 7% of S. marcescens isolates were resistant to ceftazidime, but 15% were resistant to cefotaxime. The rates of resistance to amikacin and fluoroquinolone were similar, but resistance to cefepime was lower in the ComLab isolates than in the hospital isolates.
Of P. aeruginosa hospitals isolates, 18% were resistant to piperacillin-tazobactam, 19% to ceftazidime, 21% to imipenem, 19% to amikacin, and 33% to fluoroquinolone (Table 6). The resistance rates of the ComLab P. aeruginosa isolates to piperacillin-tazobactam were similar to those of the hospital isolates, but those to ceftazidime, imipenem, amikacin, and fluoroquinolone were higher than those in the hospitals. Of the Acinetobacter spp. hospitals isolates, 26% were resistant to ampicillin-sulbactam, 19% to cefoperazone-sulbactam, and 50% to ceftazidime. The imipenem resistance rate was 22%, which was an increase from 16% in 2005. The resistance rates to amikacin and fluoroquinolone were 37% and 48%, respectively (Table 6). In the ComLab, the resistance rates to ampicillin-sulbactam, ceftazidime, and imipenem were similar, but that to amikacin was slightly lower and that to fluoroquinolone was slightly higher than those in the hospitals.
The numbers of hospitals that tested the susceptibility of non-typhoidal Salmonella were 29 in 2005 and 33 in 2007. The resistance rate of hospital isolates of this organism to ampicillin, cotrimoxazole, and fluoroquinolone was 38%, 8%, and 0%, respectively. In the ComLab, the incidence of isolates resistant to ampicillin was slightly lower (data not shown). The susceptibility of Haemophilus influenzae was tested by 18 hospitals in 2005 and 23 in 2007. Among these hospitals 19 tested both ampicillin susceptibility and β-lactamase production, 19 tested ampicillin susceptibility only, and 12 tested β-lactamase production only (data not shown). Among the 2007 isolates, 56% were resistant to ampicillin and 41% were β-lactamase positive (data not shown). The resistance rate of this organism to cotrimoxazole was 42%, which was lower than that the 56% observed in 2005.
Temporal trends of important antimicrobial agent-organism combinations in hospitals are shown in Figs. 1-4. In 2005, the proportion of MRSA, penicillin-nonsusceptible S. pneumoniae, and ampicillin-resistant E. faecium decreased slightly relative to 2003, but in 2007, the proportion of these organisms returned to the 2003 levels (Fig. 1). The proportion of vancomycin-resistant E. faecium remained unchanged from 2003 to 2005, while in 2007, the proportion was approximately five-fold higher than that ten years prior.
The rates of resistance of K. pneumoniae to ceftazidime, fluoroquinolone, and amikacin increased significantly in 2005 compared to those of previous years and then decreased slightly in 2007 (Fig. 2). Cefoxitin resistance rates of this organism remained largely unchanged since 1999. Over the last decade, fluoroquinolone resistance increased approximately four-fold, while amikacin resistance increased over two-fold.
Fewer P. aeruginosa isolates were resistant to fluoroquinolone and amikacin in 2007, but the rates of resistance to these antibiotics remained high: 33% and 19%, respectively (Fig. 3). Ceftazidime and imipenem resistance increased gradually, reaching proportions of 19% and 21%, respectively. Acinetobacter spp. showed decreased resistance to ceftazidime, fluoroquinolone, and amikacin, but the rates were still very high in 2007; 50%, 48%, and 37%, respectively. Imipenem resistance increased 22-fold over the past decade from 1% in 1997 to 22% in 2007 (Fig. 4).
Comparison of the resistance rates of antibiotic-organism combinations of concern between hospitals and the ComLab showed similar prevalences of MRSA (64% vs. 63%) and ampicillin-resistant E. faecium (86% each). Penicillin-nonsusceptible S. pneumoniae isolates were more prevalent in hospitals than in the ComLab (69% vs. 42%), vancomycin-resistant E. faecium were slightly more prevalent (21% vs. 17%), and proportions of imipenem-resistant Acinetobacter spp. were similar (22% vs. 20%). More cefotaxime-resistant E. coli (45% vs. 11%), K. pneumoniae (45% vs. 25%), and imipenem-resistant P. aeruginosa (29% vs. 21%) were detected in the isolates analyzed by the ComLab than were found in the hospital isolates.
Antimicrobial resistance rates can vary greatly among countries,16 hospitals, and even wards. For example, the European Antimicrobial Resistance Surveillance System (EARSS) Annual Report in 2008 (www.escmid.org) revealed that the proportion of MRSA was only 1% in Sweden whereas it was 53% in Portugal. In Portugal, the rate varied from 0% to 89% depending on the laboratory. This highlights the importance of a national surveillance program. To guide clinicians in the empirical selection of an appropriate antimicrobial agents, use of the first patient isolate has been recommended by the CLSI.17 However, a study showed that not all participating U.S. hospitals followed this guideline.18 Resistance selection occurring within the observation period cannot be detected when only the first isolates are included.3 In contrast to previous surveillance studies, hospitals with duplicate isolates were excluded in the present study. Inclusion of only the first isolate may result in slightly lower resistance rates, particularly for certain nosocomial pathogens.19
We found that E. coli, S. aureus, CNS, P. aeruginosa, and K. pneumoniae, in rank order, were the five most prevalent organisms in hospitals in Korea in 2007, indicating that these organisms were as important in 2007 as they were in 2004 (Table 1). In Germany, hospitals with an onsite microbiology laboratory reported higher frequencies of health care-associated infections than did hospitals without in-house laboratory services.4 This suggests that the rank order of organisms in the ComLab may not be accurate.
The most prevalent organisms isolated from ICU patients in 2000-2002 were S. aureus in the US, Canada, and France, P. aeruginosa in Italy, and CNS in Germany.16 A 2006-2007 National Healthcare Safety Network (NHSN, which includes the former National Nosocomial Infections Surveillance) reported that the three most prevalent organisms isolated from healthcare-associated infections (HAI) were CNS (15%), S. aureus (15%), and Enterococcus sp. (12%).20 This suggests that our isolates were mostly from inpatients and hence had high resistance rates. A. baumannii can easily acquire antibiotic resistance determinants.21 We therefore predicted that this organism would become more prevalent, but it became less prevalent in the hospitals, with a change in rank from the sixth position in 2004 to the eighth position in 2007. In the ComLab data, the typical nosocomial nonfermentative pathogen P. aeruginosa ranked second in 2007, as in 2004.6
The CLSI document17 recommends groups of antimicrobial agents for susceptibility testing. Group A agents are recommended for primary testing and reporting, while Group B contains agents that may warrant primary testing, however, they should be reported only selectively, such as when the organism is resistant to agents of the same family. However, these recommendations are difficult to follow in practice when resistant organisms are very prevalent. When a laboratory tests only a few classes of antimicrobial agents, information on other agents of interest to clinicians is not available, resulting in potential misselection of antimicrobial agents. More laboratories used cephalothin, piperacillin- tazobactam, and cefoxitin to test E. coli in 2007 than in 2002, but still the laboratories using ampicillin-sulbactam (or amoxicillin-clavulanic acid) and cefoxitin were only 65% and 81%, respectively. Only 51% and 61% of laboratories used cotrimoxazole to test for E. coli and S. aureus, respectively.
Certain antibiotic-organism combinations are relatively more prevalent in Korea and other Asian countries than in the rest of the world. The results of the present surveillance study highlight the fact that MRSA continues to be prevalent in Korea, with an occurrence rate of 64% (Fig. 1). In a nationwide surveillance study in Japan in 2007, 59.7% of respiratory isolates of S. aureus were methicillin-resistant.22 Analysis of data from the Surveillance Network-USA (TSN), an electronic surveillance network that collected microbiology data from 300 clinical microbiology laboratories across the US, revealed that the proportions of MRSA in 2005 in non-ICU inpatients, ICU patients, and outpatients were 59.2%, 55%, and 47.9%, respectively.23 All of the reports discussed above indicate that MRSA has also become a serious problem in non-Asian countries.
The CLSI document17 defined two different penicillin G susceptibility breakpoints for S. pneumoniae depending on the site of isolation. The vast majority of S. pneumoniae isolates are from non-meningitis patients. Therefore, applying meningitis breakpoint to all S. pneumoniae appears illogical, but in epidemiologic surveillance studies, it was suggested that continued reporting based on meningitis (≤ 0.06 µg/mL) breakpoint might also be useful to allow for continuity in the reporting of long-term resistance occurrence data.24 These investigators reported that the MIC range of penicillin for a large number of worldwide isolates in 2006 was ≤ 0.03-32 µg/mL, but 68% of the isolates were inhibited by ≤ 0.06 µg/mL, 93% by 2 µg/mL (non-meningitis susceptible), and 99% by 4 µg/mL (non-meningitis intermediate). In our previous surveillance study, penicillin susceptibility testing was performed mostly using the oxacillin disk test to screen nonsusceptible isolates based on the meningitis breakpoint. In the current study, penicillin-nonsusceptible S. pneumoniae remained very prevalent (69%) in hospitals in 2007 (Fig. 5), but the rate in the ComLab was much lower (42%) (Table 3). A high prevalence of penicillin-nonsusceptible S. pneumoniae has also been reported in other Asian countries.25
The resistance rates of E. faecalis to ampicillin and vancomycin are very low. However, since 2003, the vancomycin resistance rate of E. faecium has been reported to be 20% or higher in the hospitals (Fig. 5). The ComLab rates of E. faecium vancomycin-resistance were 12% in 2005 and 17% in 2007 (Table 3); a significant increase compared to 7% in 2003.26 Vancomycin-resistant E. faecium has also become more prevalent in the United States. In a Chicago Hospital, the prevalence of vancomycin-resistant E. faecium increased from 28.9% in 1993 to 72.4% in 2002, based on one isolate per patient per month.27 The NHSN report20 stated that among the isolates from catheterassociated urinary tract infections and central line-associated bloodstream infections in 2006 and 2007, 90.4% and 80% of E. faecium were resistant to ampicillin and to vancomycin, respectively. This indicates that vancomycin-resistant enterococci are difficult to control. The rates of E. faecium resistance to ampicillin were similar in the current study and in the NHSN report, but vancomycin-resistance was lower in our study than in the NHSN report.
The prevalence of ampicillin-resistant E. coli in hospitals was 60% in 2007, a slight decrease compared to the 2004 rate, but in the ComLab it was 70%. This finding indicates that empirical selection for ampicillin is difficult, but that a significant proportion of E. coli infections can be treated using ampicillin after laboratory testing. E. coli and K. pneumoniae often acquire ESBL genes. In 2007, 11% of E. coli and 29% of K. pneumoniae hospital isolates were resistant to cefotaxime or ceftazidime, respectively, suggesting ESBL production. Some hospitals tested E. coli and K. pneumoniae for both susceptibility to third-generation cephalosporins and the production of β-lactamases. The resistance rates to third-generation cephalosporins and ESBL positive rates in 2007 were E. coli 12% and 12%, respectively, and K. pneumoniae 32% and 27%, respectively. It is of concern that in the ComLab in 2007, the ceftazidime-resistance rates of both E. coli and K. pneumoniae were much higher (47%) than those documented for hospitals. An outbreak of SHV-12 type ESBL-producing K. pneumoniae was recently reported in a Korean hospital.28
In the hospitals, 6% of E. coli and 21% of K. pneumoniae were resistant to cefoxitin, suggesting plasmid-mediated AmpC production. A previous study showed that the high prevalence of cefoxitin-resistant K. pneumoniae isolates was due to the plasmid-mediated AmpC enzymes, DHA-1 and CMY-1.10 It was reported that among the isolates from the Asia-Pacific region in 1998-2004, 8.9% of E. coli and 20.3% of K. pneumoniae isolates were ESBL screen-positive, but significant proportions of them were confirmation test negative.29 Among these nonconfirming isolates, 62% of E. coli and 75% of K. pneumoniae harbored a plasmid-borne AmpC enzyme of the CIT or DHA type. The high cefoxitin resistance rates of E. coli and K. pneumoniae in our present study, 6% and 21%, respectively, indicate that ESBL detection could be difficult, resulting in underestimation of the prevalence.
The percentages of ceftazidime-resistant E. cloacae and S. marcescens isolates from the hospitals in 2007 were 30% and 17%, respectively; these values were slightly lower than those reported in 2004, namely 35% and 23%, respectively. The resistance rates to cefepime remained low, and imipenem-resistant Enterobacteriaceae remained very rare, but KPC type class A carbapenemase may emerge in the near future in Korea. K. pneumoniae isolates producing KPC carbapenemase are commonly isolated in health care institutions located in the Northeast US, but the resistance has also spread to other parts of the country,30 as well as to other counties, including China.31
The rates of ampicillin resistance and β-lactamase production in H. influenzae were similar in 2004 (47% and 51%, respectively), but in 2007, the rates were 51% and 41%, respectively. It is necessary to determine whether this difference is indeed due to the emergence of β-lactamase-negative ampicillin-resistant (BNAR) H. influenzae, which have become prevalent in Japan.32 The resistance rates of non-typhoidal Salmonella to cotrimoxazole (8%) and fluoroquinolone (0%) in hospitals were similar to those in 2004, but the ampicillin resistance rate decreased slightly from 44% to 38%. Fluoroquinolone resistance rate, despite being as low as 0%, probably fails to indicate clinical efficacy in the treatment of extraintestinal infections. This is because low-level quinolone resistance cannot be detected using fluoroquinolones.17
P. aeruginosa and Acinetobacter spp. are important nosocomial pathogens. With the emergence of multidrug-resistant isolates, few drugs are now available to treat Acinetobacter infections. Polymyxins are the only therapeutic option in many cases,33 but the emergence of A. baumannii isolates resistant to all tested antimicrobials, including polymyxin B, have been reported in Korea.34 Fewer isolates of P. aeruginosa and Acinetobacter spp. were resistant to fluoroquinolone and amikacin in our study than has been documented previously. The rates of imipenem resistance in P. aeruginosa remained stable, but more Acinetobacter spp. isolates were impenem-resistant than has been reported previously. It was shown that in P. aeruginosa, only 10.8% of imipenem resistance was due to metallo-β-lactamase production,8 but the majority of resistance in Acinetobacter spp. was due to OXA-type carbapenemase production.9 Outbreak of imipenem-resistant A. baumannii with OXA-23 carbapenemase was recently reported in a Korean tertiary care hospital.35 Increasing carbapenem resistance is a serious problem, because this is the only class of β-lactam agents active against isolates producing ESBLs or derepressed AmpC enzymes.36
In contrast to our predictions, third-generation cephalosporin-resistant E. coli and K. pneumoniae strains and imipenem-resistant P. aeruginosa strains were more prevalent in isolates analyzed by the ComLab (Fig. 5) than in isolates examined by the hospital laboratory, suggesting that antimicrobial-resistant nosocomial pathogens are also a serious problem at smaller hospitals and clinics that have no in-house microbiology laboratory service.
In conclusion, MRSA, third-generation cephalosporin-resistant K. pneumoniae, and fluoroquinolone-resistant E. coli, P. aeruginosa and Acinetobacter spp. are still prevalent in the hospitals, and the smaller hospitals and clinics in Korea, and vancomycin-resistant E. faecium, and imipenem-resistant Acinetobacter spp. have gradually increased in Korean hospitals over the last decade. The higher prevalences of third-generation cephalosporin-resistant E. coli and K. pneumoniae and imipenem-resistant P. aeruginosa in ComLab isolates are of concern.
Figures and Tables
 | Fig. 1The resistance trends of gram-positive cocci in the hospitals. Oxacillin-resistant S. aureus, penicillin-nonsusceptible S. pneumoniae, and ampicillin-resistant E. faecium continued to have high prevalence rates, and a gradual increase in vancomycin-resistant E. faecium strains was observed. OXA, oxacillin; PEN, penicillin; AMP, ampicillin; VAN, vancomycin; R, resistant; NS, nonsusceptible; SAU, S. aureus; SPN, S. pneumoniae; EFM, E. faecium. |
 | Fig. 2The resistance trends of K. pneumoniae in the hospitals. The cefoxitin resistance rate remained high but stable, whereas ceftazidime, fluoroquinolone, and amikacin resistance rates increased in 2005 compared to those of previous years. CAZ, ceftazidime; FOX, cefoxitin; FQN, fluoroquinolone; AMK, amikacin. |
 | Fig. 3The resistance trends of P. aeruginosa isolates from hospitals. The resistance rates to fluoroquinolone and amikacin declined slightly, but those to ceftazidime and imipenem remained stable. FQN, fluoroquinolone; AMK, amikacin; CAZ, ceftazidime; IPM, imipenem. |
 | Fig. 4The resistance trends of Acinetobacter spp. isolates in hospitals. The resistance rates to ceftazidime, fluoroquinolone, and amikacin decreased slightly, but that to imipenem increased steadily. FQN, fluoroquinolone; AMK, amikacin; CAZ, ceftazidime; IPM, imipenem. |
 | Fig. 5Comparison of the prevalence of antimicrobial-organism combinations in the hospitals and the commercial laboratory (ComLab) in 2007. Penicillin-nonsusceptible S. pneumoniae and vancomycin-resistant E. faecium were more prevalent in the hospitals. Cefotaxime-resistant E. coli and K. pneumoniae and imipenem-resistant P. aeruginosa were more prevalent among isolates in the commercial laboratory. OXA, oxacillin; R, resistant; SAU, S. aureus; PEN, penicillin; NS, nonsusceptible; SPN, S. pneumoniae; AMP, ampicillin; EFM, E. faecium; VAN, vancomycin; CTX, cefotaxime; ECO, E. coli; KPN, K. pneumoniae; IPM, imipenem; ACI, Acinetobacter spp.; PAE, P. aeruginosa. |
Table 1
Comparison of the Number, Proportion, and Rank Order of Organisms Detected in Surveillance Programs in 1998, 2004, and 2007
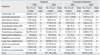
Table 2
Antimicrobial Resistant Rates (%) of S. aureus and Coagulase-Negative Staphylococcus Isolated in 2005 and in 2007

Table 3
Antimicrobial Resistant Rates (%) of S. pneumoniae, E. faecalis, and E. faecium Isolated in 2005 and in 2007
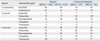
Table 4
Antimicrobial Resistance Rates (%) of E. coli and K. pneumoniae Isolated in 2005 and in 2007

ACKNOWLEDGEMENTS
Other KONSAR program participants
Nam Hee Ryoo, Keimyong University Dong San Medical Center, Daegu; Seok Hoon Jeong, Kosin University Gospel Hospital, Busan; Gyoung Yim Ha, Dongguk University Kyongju Hospital, Gyeongju; Mi-Na Kim, University of Ulsan College of Medicine, Seoul; Wee Gyo Lee, Ajou University Hospital, Suwon; Young Ae Hong, Ulsan Dong-Kang General Hospital, Ulsan; Sung Eun Cho, Ewha Womans University College of Medicine, Seoul; Young Uh, Yonsei University Wonju Christian Hospital, Wonju; Sook Jin Jang, Chosun University Hospital, Gwangju; Myung Hee Lee, Korea Veterans Hospital, Seoul; Wonkeun Song, Hallym University College of Medicine, Seoul; Tae Yeal Choi, Hanyang University Medical College, Seoul; Jong-Hee Shin, Chonnam National University Hospital, Gwangju; Seong Geun Hong, Pochon CHA University Hospital, Seongnam; Young Ah Kim, National Health Insurance Corporation Ilsan Hospital, Goyang; Dong-Hee Cho, Samsung Cheil Hospital, Seoul; Dong Hee Whang, Inje University College of Medicine, Seoul; Seungok Lee, Seoul Clinical Laboratory, Seoul; Seong Hee Lee, Hanmaeum Hospital, Jeju.
References
1. Jones RN, Masterton R. Determining the value of antimicrobial surveillance programs. Diagn Microbiol Infect Dis. 2001. 41:171–175.

2. Morris AK, Masterton RG. Antibiotic resistance surveillance: action for international studies. J Antimicrob Chemother. 2002. 49:7–10.

3. Cornaglia G, Hryniewicz W, Jarlier V, Kahlmeter G, Mittermayer H, Stratchounski L, et al. European recommendations for antimicrobial resistance surveillance. Clin Microbiol Infect. 2004. 10:349–383.

4. Llata E, Gaynes RP, Fridkin S. Measuring the scope and magnitude of hospital-associated infection in the United States: the value of prevalence surveys. Clin Infect Dis. 2009. 48:1434–1440.

5. Chong Y, Lee K, Park YJ, Jeon DS, Lee MH, Kim MY, et al. Korean Nationwide Surveillance of Antimicrobial Resistance of bacteria in 1997. Yonsei Med J. 1998. 39:569–577.

6. Lee K, Lim CH, Cho JH, Lee WG, Uh Y, Kim HJ, et al. High prevalence of ceftazidime-resistant Klebsiella pneumoniae and increase of imipenem-resistant Pseudomonas aeruginosa and Acinetobacter spp. in Korea: a KONSAR program in 2004. Yonsei Med J. 2006. 47:634–645.

7. Lee K, Lee WG, Uh Y, Ha GY, Cho J, Chong Y. Korean Nationwide Surveillance of Antimicrobial Resistance Group. VIM- and IMP-type metallo-beta-lactamase-producing Pseudomonas spp. and Acinetobacter spp. in Korean hospitals. Emerg Infect Dis. 2003. 9:868–871.

8. Lee K, Park AJ, Kim MY, Lee HJ, Cho JH, Kang JO, et al. Metallo-beta-lactamase-producing Pseudomonas spp. in Korea: high prevalence of isolates with VIM-2 type and emergence of isolates with IMP-1 type. Yonsei Med J. 2009. 50:335–339.

9. Lee K, Kim MN, Choi TY, Cho SE, Lee S, Whang DH, et al. Wide dissemination of OXA-type carbapenemases in clinical Acinetobacter spp. isolates from South Korea. Int J Antimicrob Agents. 2009. 33:520–524.
10. Lee K, Lee M, Shin JH, Lee MH, Kang SH, Park AJ, et al. Prevalence of plasmid-mediated AmpC beta-lactamases in Escherichia coli and Klebsiella pneumoniae in Korea. Microb Drug Resist. 2006. 12:44–49.

11. Fridkin SK, Hill HA, Volova NV, Edwards JR, Lawton RM, Gaynes RP, et al. Temporal changes in prevalence of antimicrobial resistance in 23 US hospitals. Emerg Infect Dis. 2002. 8:697–701.
12. Van Beneden CA, Lexau C, Baughman W, Barnes B, Bennett N, Cassidy PM, et al. Aggregated antibiograms and monitoring of drug-resistant Streptococcus pneumoniae. Emerg Infect Dis. 2003. 9:1089–1095.
13. Bax R, Bywater R, Cornaglia G, Goosens H, Hunter P, Isham V, et al. Surveillance of antimicrobial resistance-what, how and whither? Clin Microbiol Infect. 2001. 7:316–325.
14. Sahm DF, Marsilio MK, Piazza G. Antimicrobial resistance in key bloodstream bacterial isolates: electronic surveillance with the Surveillance Network Database-USA. Clin Infect Dis. 1999. 29:259–263.

15. Felmingham D, Grüeneberg RN. The Alexander project 1996-1997: latest susceptibility data from this international study of bacterial pathogens from community-acquired lower respiratory tract infections. J Antimicrob Chemother. 2000. 45:191–203.
16. Jones ME, Draghi DC, Thornsberry C, Karlowsky JA, Sahm DF, Wenzel RP. Emerging resistance among bacterial pathogens in the intensive care unit - a European and North American Surveillance study (2000-2002). Ann Clin Microbiol Antimicrob. 2004. 3:14.
17. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; nineteenth informational supplement, M100-S19. 2009. Wayne, PA: CLSI.
18. Halstead DC, Gomez N, McCarter YS. Reality of developing a community-wide antibiogram. J Clin Microbiol. 2004. 42:1–6.

19. Lee SO, Cho YK, Kim SY, Lee ES, Park SY, Seo YH. Comparison of trends of resistance rates over 3 years calculated from results for all isolates and for the first isolate of a given species from a patient. J Clin Microbiol. 2004. 42:4776–4779.

20. Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect Control Hosp Epidemiol. 2008. 29:996–1011.

21. Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008. 21:538–582.

22. Niki Y, Hanaki H, Matsumoto T, Yagisawa M, Kohno S, Aoki N, et al. Nationwide surveillance of bacterial respiratory pathogens conducted by the Japanese Society of Chemotherapy in 2007: general view of the pathogens' antibacterial susceptibility. J Infect Chemother. 2009. 15:156–167.
23. Styers D, Sheehan DJ, Hogan P, Sahm DF. Laboratory-based surveillance of current antimicrobial resistance patterns and trends among Staphylococcus aureus: 2005 status in the United States. Ann Clin Microbiol Antimicrob. 2006. 5:2.
24. Weinstein MP, Klugman KP, Jones RN. Rationale for revised penicillin susceptibility breakpoints versus Streptococcus pneumoniae: coping with antimicrobial susceptibility in an era of resistance. Clin Infect Dis. 2009. 48:1596–1600.

25. Song JH, Jung SI, Ko KS, Kim NY, Son JS, Chang HH, et al. High prevalence of antimicrobial resistance among clinical Streptococcus pneumoniae isolates in Asia (an ANSORP study). Antimicrob Agents Chemother. 2004. 48:2101–2107.

26. Lee K, Park KH, Jeong SH, Lim HS, Shin JH, Yong D, et al. Further increase of vancomycin-resistant Enterococcus faecium, amikacin- and fluoroquinolone-resistant Klebsiella pneumoniae, and imipenem-resistant Acinetobacter spp. in Korea: 2003 KONSAR surveillance. Yonsei Med J. 2006. 47:43–54.
27. Treitman AN, Yarnold PR, Warren J, Noskin GA. Emerging incidence of Enterococcus faecium among hospital isolates (1993 to 2002). J Clin Microbiol. 2005. 43:462–463.
28. Roh KH, Uh Y, Kim JS, Kim HS, Shin DH, Song W. First outbreak of multidrug-resistant Klebsiella pneumoniae producing both SHV-12-type extended-spectrum beta-lactamase and DHA-1-type AmpC beta-lactamase at a Korean hospital. Yonsei Med J. 2008. 49:53–57.

29. Bell JM, Chitsaz M, Turnidge JD, Barton M, Walters LJ, Jones RN. Prevalence and significance of a negative extended-spectrum beta-lactamase (ESBL) confirmation test result after a positive ESBL screening test result for isolates of Escherichia coli and Klebsiella pneumoniae: results from the SENTRY Asia-Pacific Surveillance Program. J Clin Microbiol. 2007. 45:1478–1482.

30. Kitchel B, Sundin DR, Patel JB. Regional dissemination of KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother. 2009. 53:4511–4513.
31. Cai JC, Zhou HW, Zhang R, Chen GX. Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob Agents Chemother. 2008. 52:2014–2018.

32. Hasegawa K, Chiba N, Kobayashi R, Murayama Y, Iwata S, Sunakawa K, et al. Rapidly increasing prevalence of beta-lactamase-nonproducing, ampicillin-resistant Haemophilus influenzae type b in patients with meningitis. Antimicrob Agents Chemother. 2004. 48:1509–1514.

33. Walkty A, DeCorby M, Nichol K, Karlowsky JA, Hoban DJ, Zhanel GG. In vitro activity of colistin (polymyxin E) against 3,480 isolates of gram-negative bacilli obtained from patients in Canadian hospitals in the CANWARD Study, 2007-2008. Antimicrob Agents Chemother. 2009. 53:4924–4926.

34. Park YK, Peck KR, Cheong HS, Chung DR, Song JH, Ko KS. Extreme drug resistance in Acinetobacter baumannii infections in intensive care units, South Korea. Emerg Infect Dis. 2009. 15:1325–1327.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download